Summary information and primary citation
- PDB-id
- 484d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
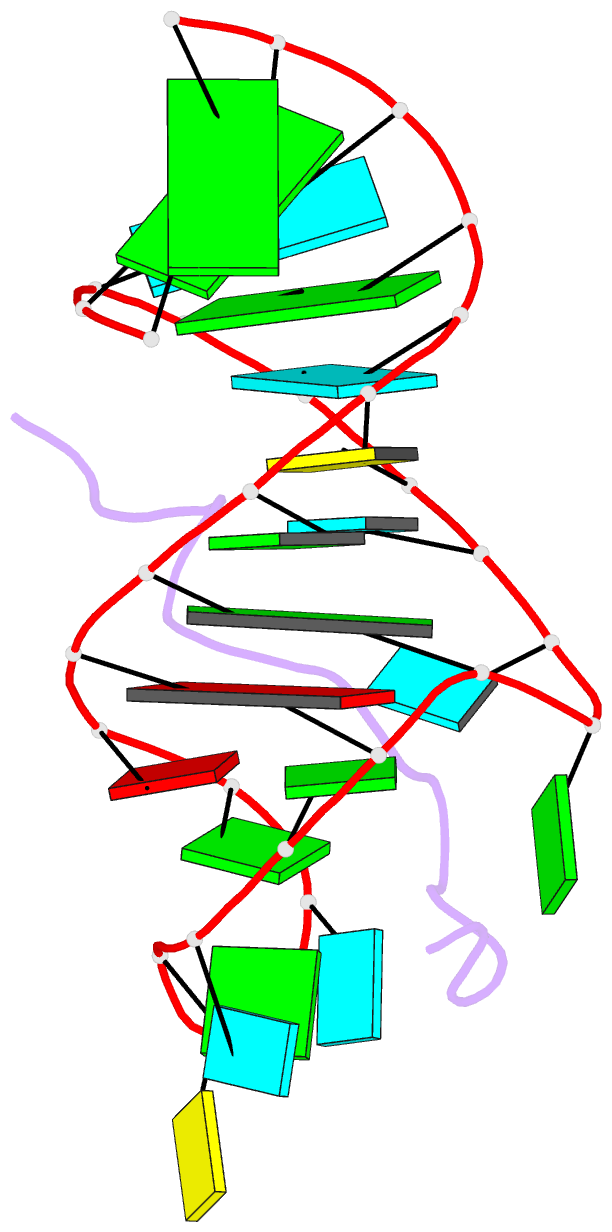
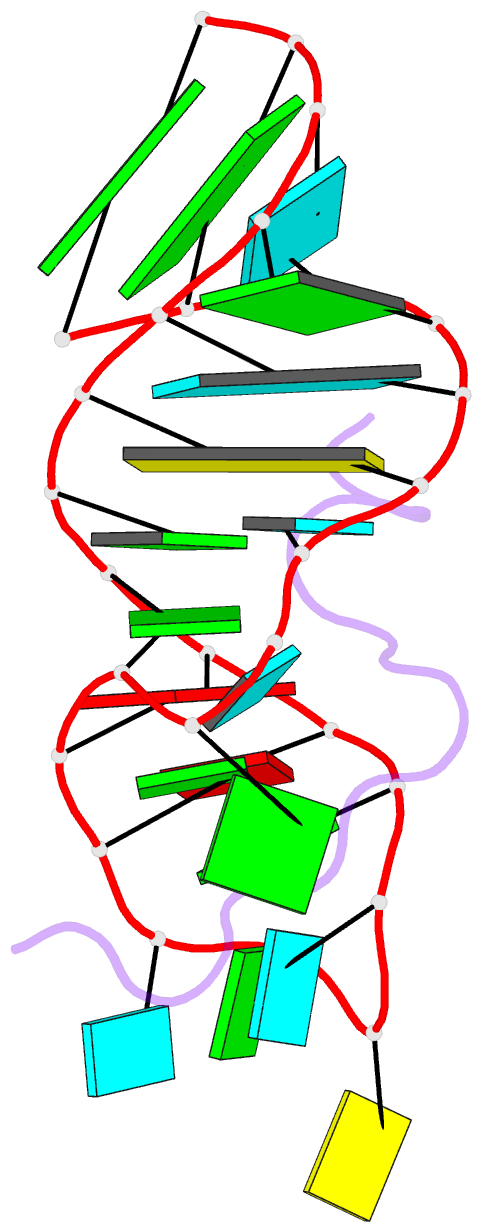
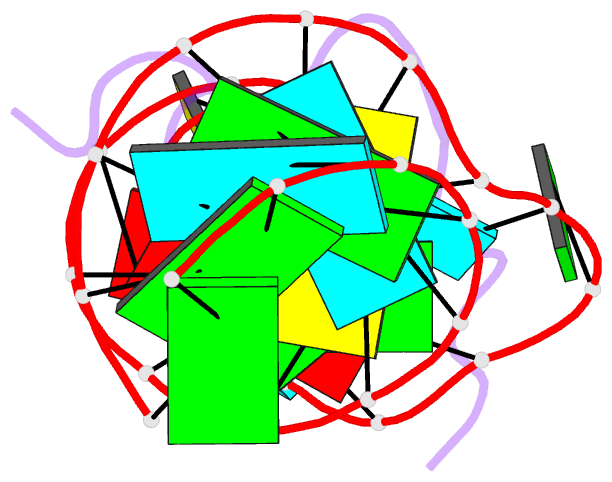
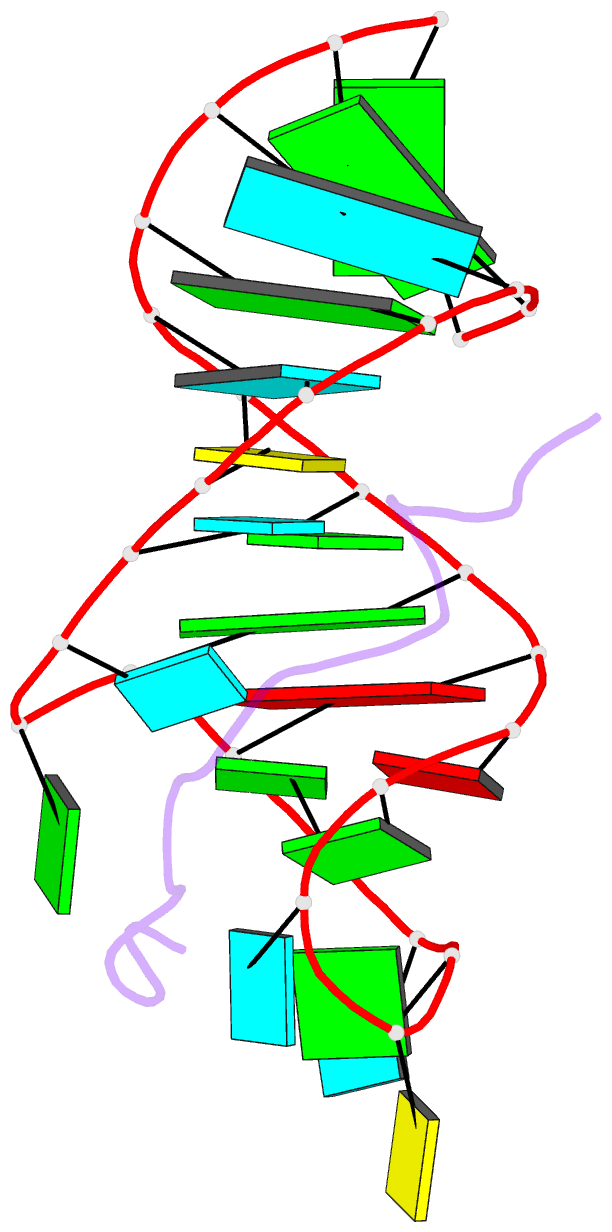
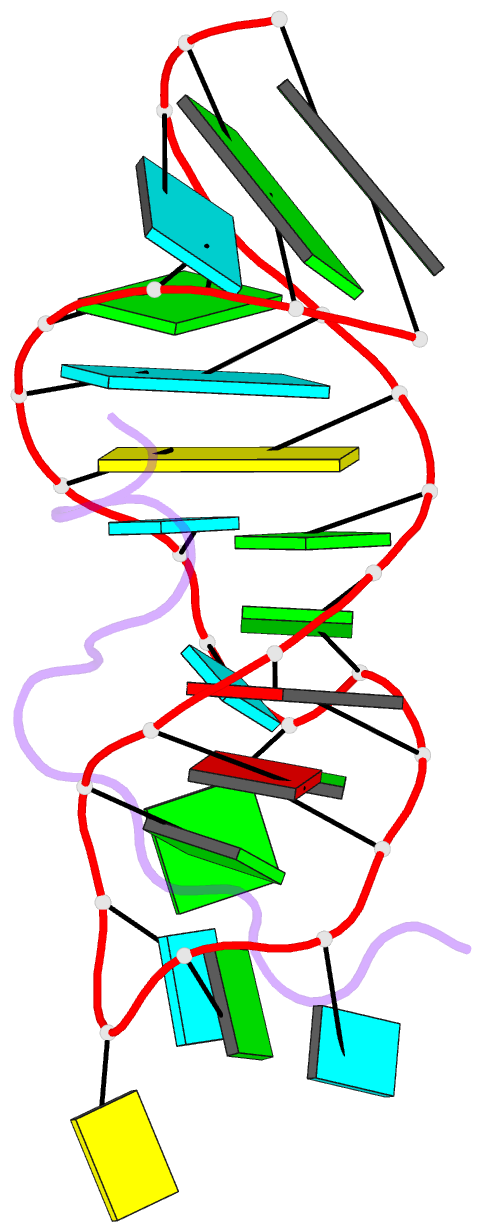
- Solution structure of hiv-1 rev peptide-RNA aptamer complex
- Reference
- Ye X, Gorin A, Frederick R, Hu W, Majumdar A, Xu W, McLendon G, Ellington A, Patel DJ (1999): "RNA architecture dictates the conformations of a bound peptide." Chem.Biol., 6, 657-669. doi: 10.1016/S1074-5521(99)80117-3.
- Abstract
- Background: The biological function of several viral and bacteriophage proteins, and their arginine-rich subdomains, involves RNA-mediated interactions. It has been shown recently that bound peptides adopt either beta-hairpin or alpha-helical conformations in viral and phage peptide-RNA complexes. We have compared the structures of the arginine-rich peptide domain of HIV-1 Rev bound to two RNA aptamers to determine whether RNA architecture can dictate the conformations of a bound peptide.
Results: The core-binding segment of the HIV-1 Rev peptide class II RNA aptamer complex spans the two-base bulge and hairpin loop of the bound RNA and the carboxy-terminal segment of the bound peptide. The bound peptide is anchored in place by backbone and sidechain intermolecular hydrogen bonding and van der Waals stacking interactions. One of the bulge bases participates in U*(A*U) base triple formation, whereas the other is looped out and flaps over the bound peptide in the complex. The seven-residue hairpin loop is closed by a sheared G*A mismatch pair with several pyrimidines looped out of the hairpin fold.
Conclusions: Our structural studies establish that RNA architecture dictates whether the same HIV-1 Rev peptide folds into an extended or alpha-helical conformation on complex formation. Arginine-rich peptides can therefore adapt distinct secondary folds to complement the tertiary folds of their RNA targets. This contrasts with protein-RNA complexes in which elements of RNA secondary structure adapt to fit within the tertiary folds of their protein targets.