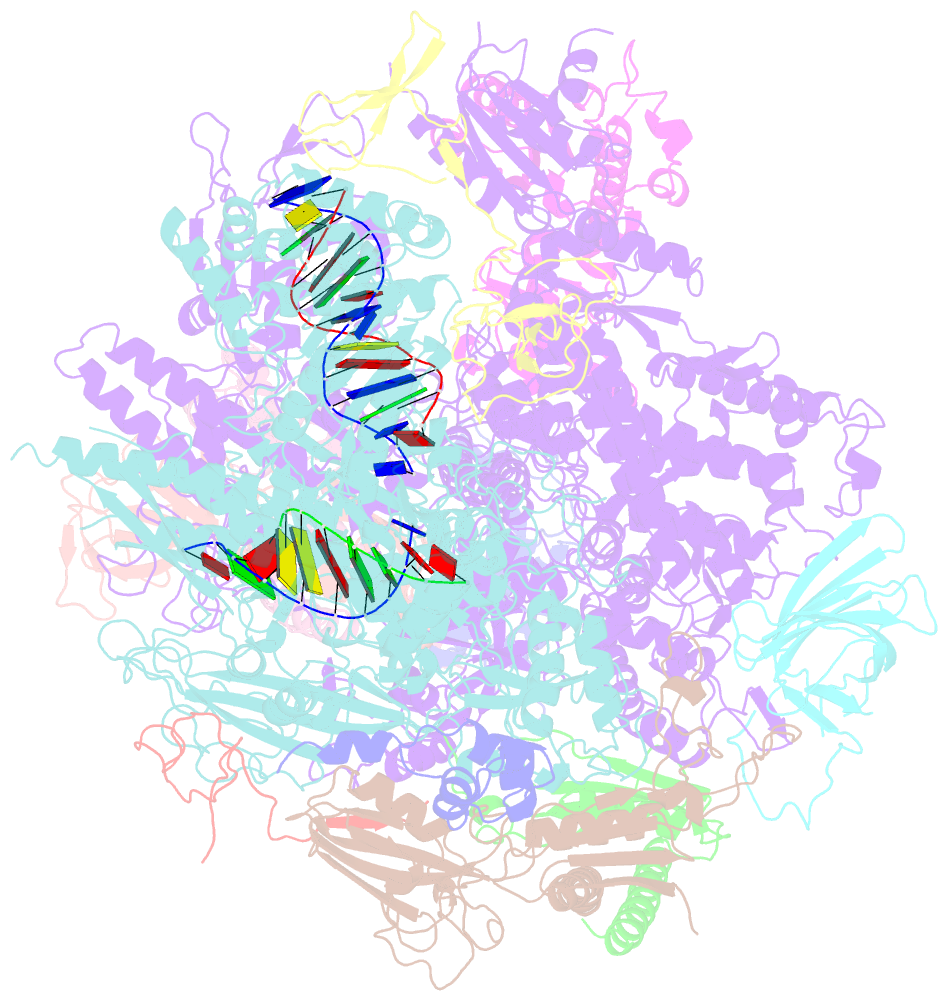
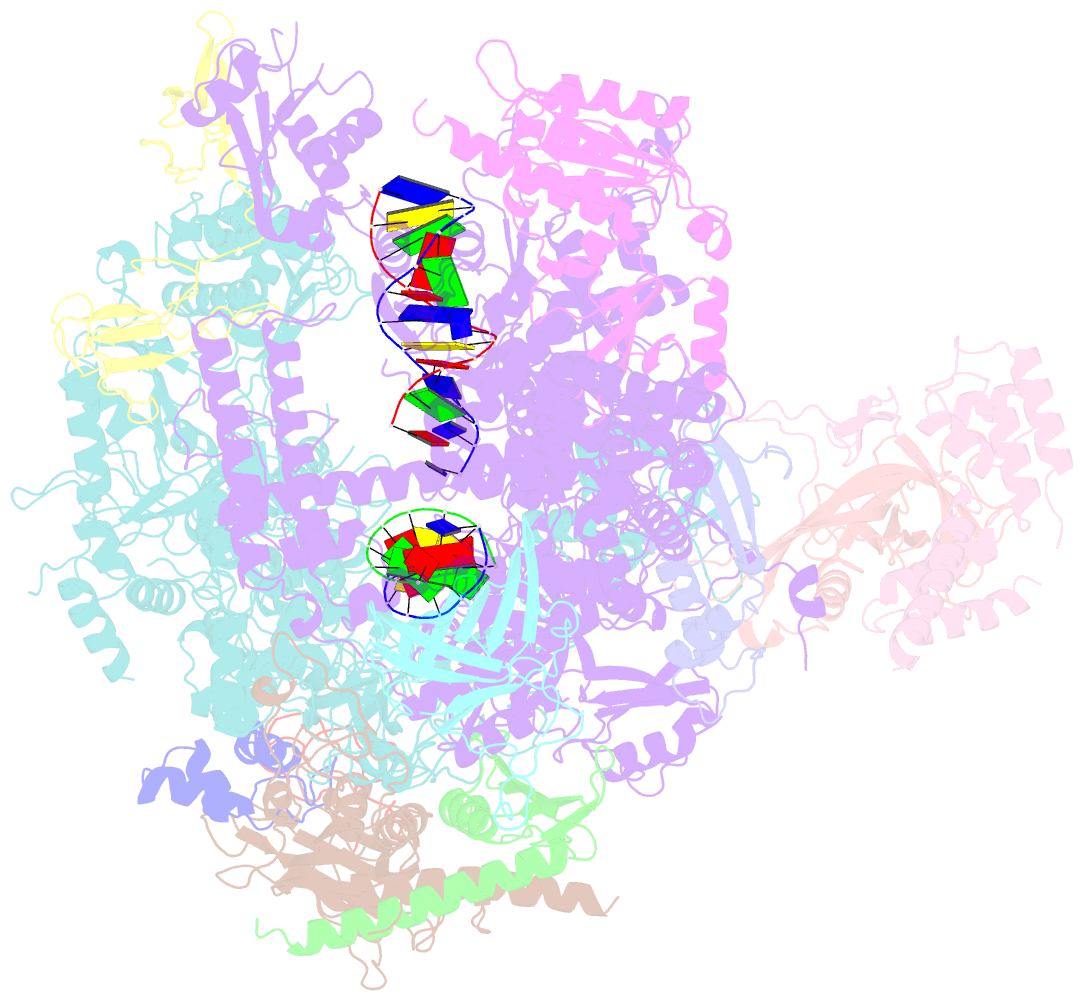
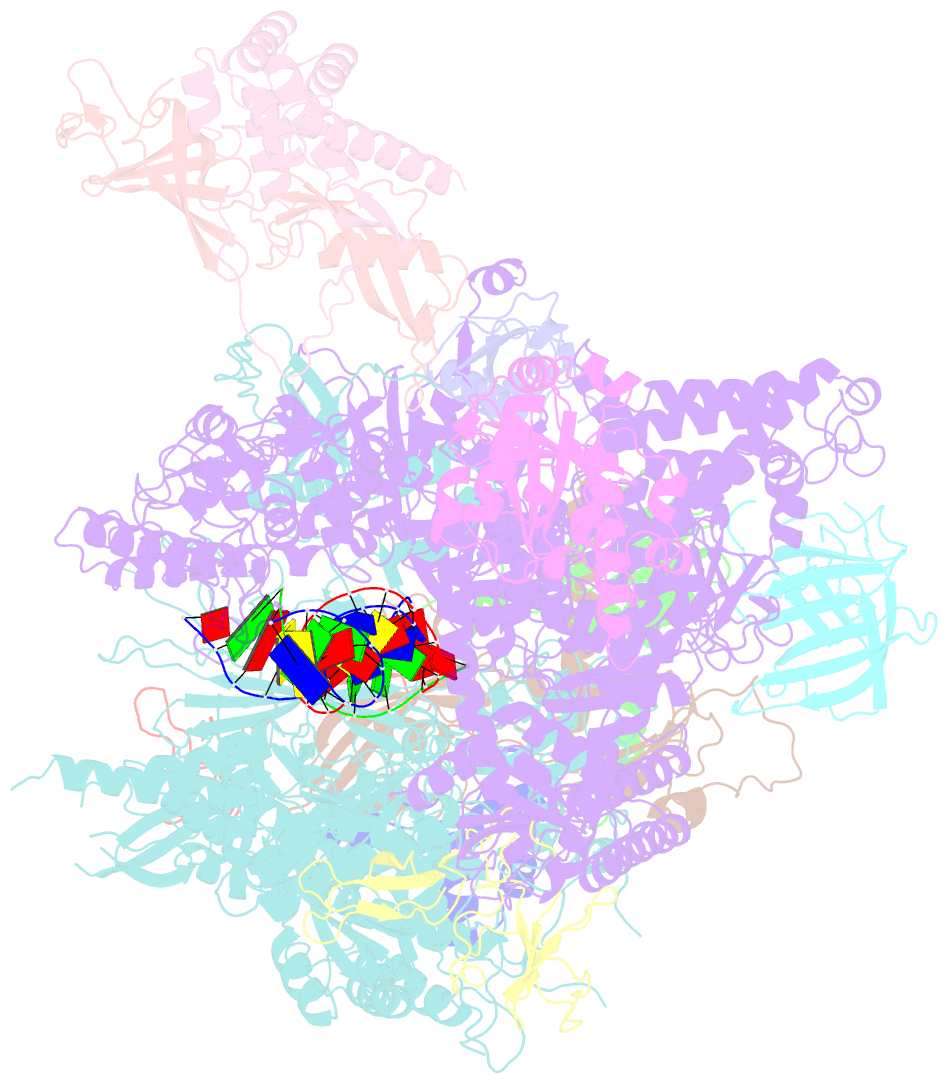
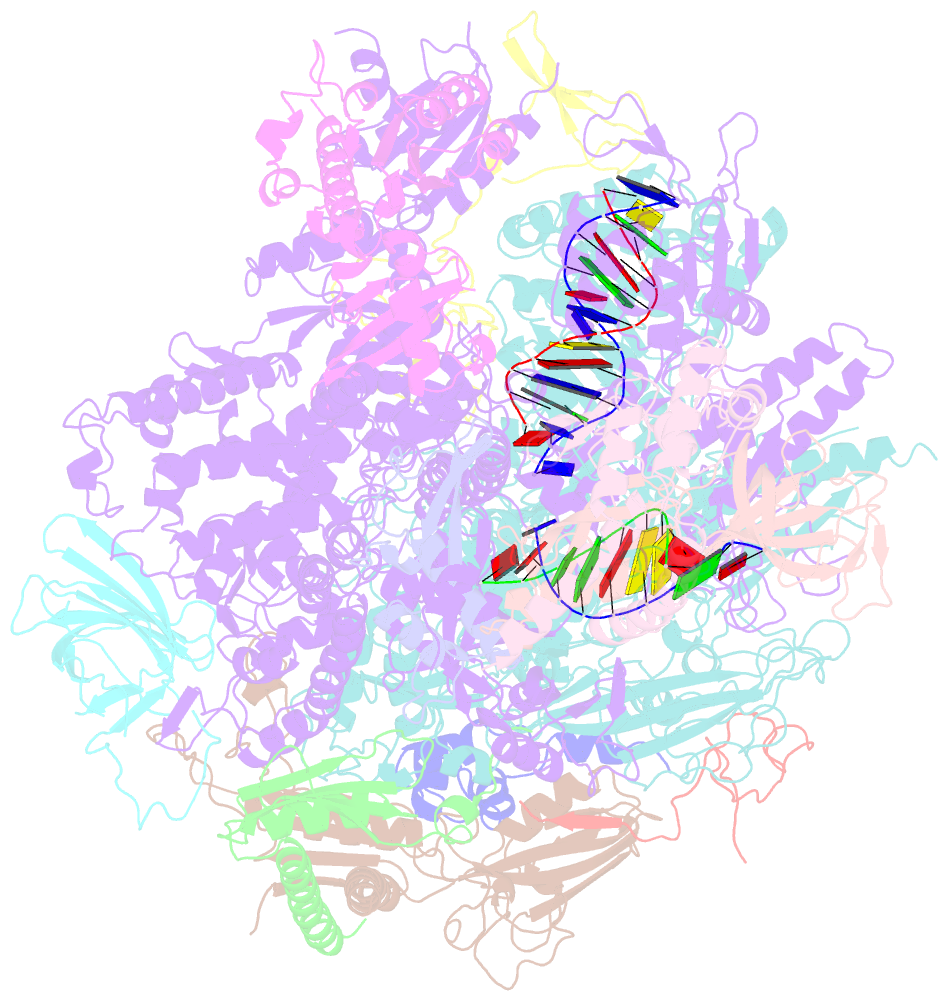
Summary information and primary citation
- PDB-id
- 4a93; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (3.4 Å)
- Summary
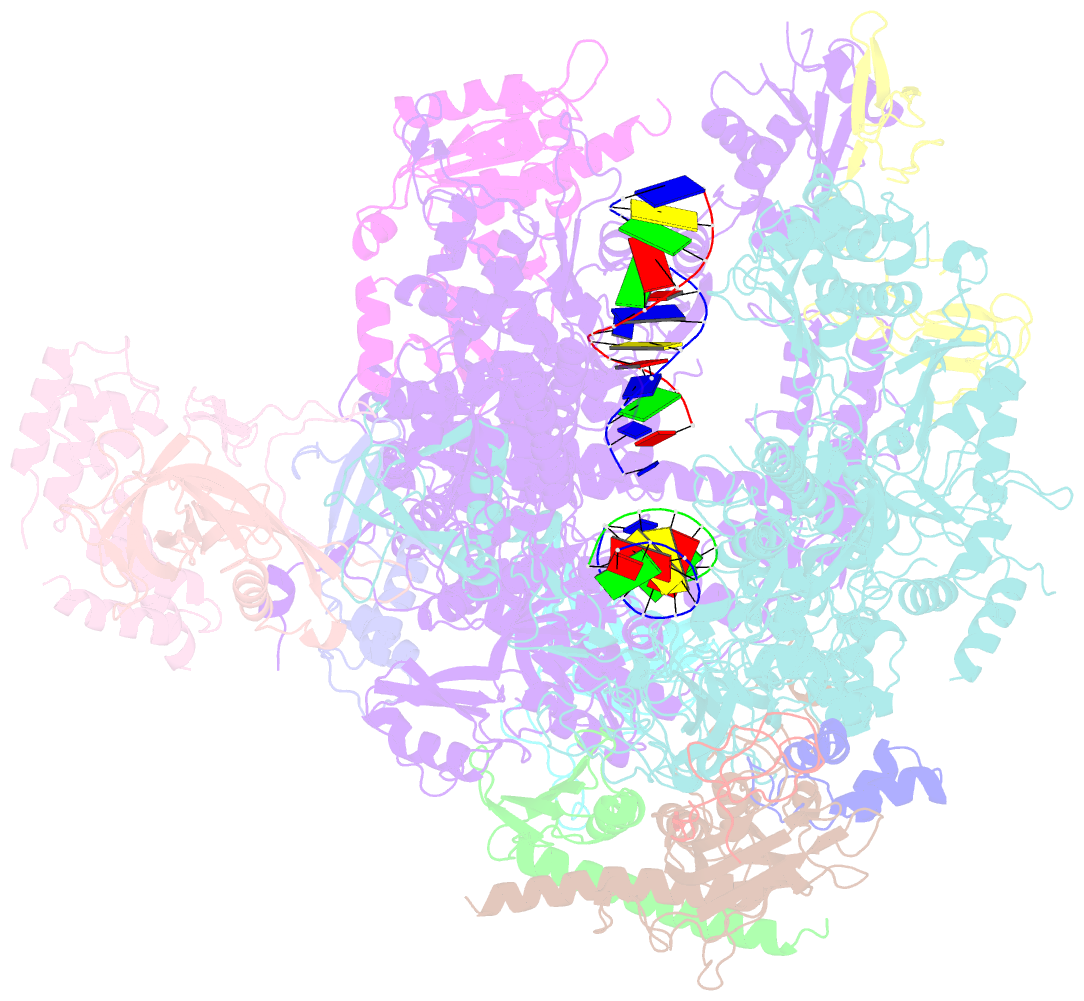
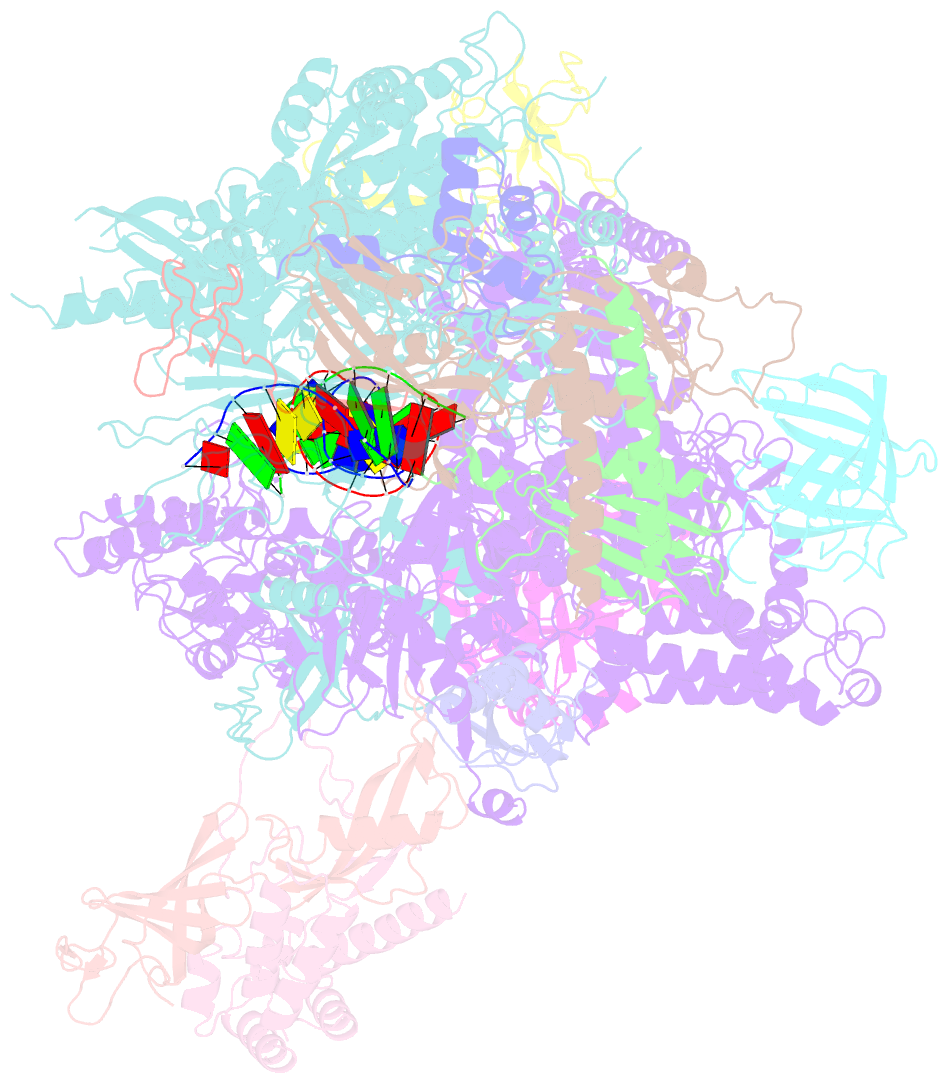
- RNA polymerase ii elongation complex containing a cpd lesion
- Reference
- Walmacq C, Cheung AC, Kireeva ML, Lubkowska L, Ye C, Gotte D, Strathern JN, Carell T, Cramer P, Kashlev M (2012): "Mechanism of Translesion Transcription by RNA Polymerase II and its Role in Cellular Resistance to DNA Damage." Mol.Cell, 46, 18. doi: 10.1016/J.MOLCEL.2012.02.006.
- Abstract
- UV-induced cyclobutane pyrimidine dimers (CPDs) in the template DNA strand stall transcription elongation by RNA polymerase II (Pol II). If the nucleotide excision repair machinery does not promptly remove the CPDs, stalled Pol II creates a roadblock for DNA replication and subsequent rounds of transcription. Here we present evidence that Pol II has an intrinsic capacity for translesion synthesis (TLS) that enables bypass of the CPD with or without repair. Translesion synthesis depends on the trigger loop and bridge helix, the two flexible regions of the Pol II subunit Rpb1 that participate in substrate binding, catalysis, and translocation. Substitutions in Rpb1 that promote lesion bypass in vitro increase UV resistance in vivo, and substitutions that inhibit lesion bypass decrease cell survival after UV irradiation. Thus, translesion transcription becomes essential for cell survival upon accumulation of the unrepaired CPD lesions in genomic DNA.