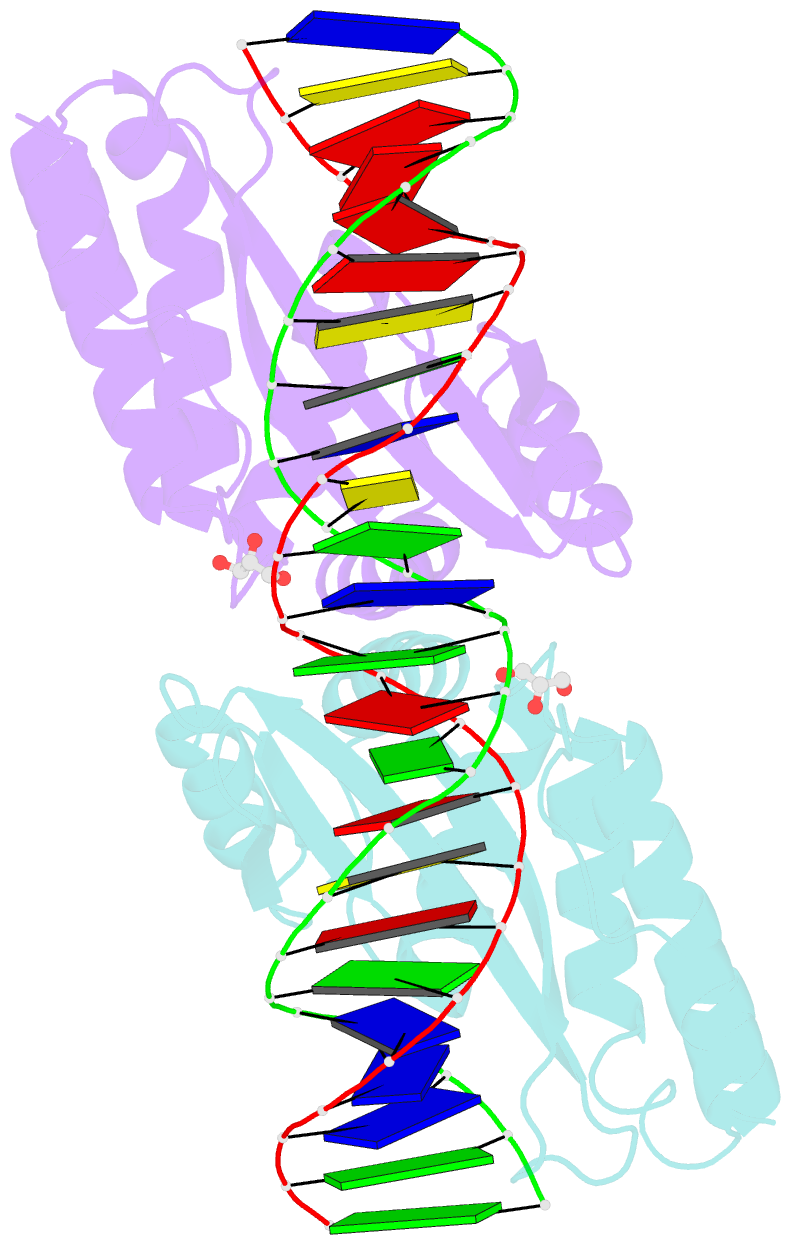
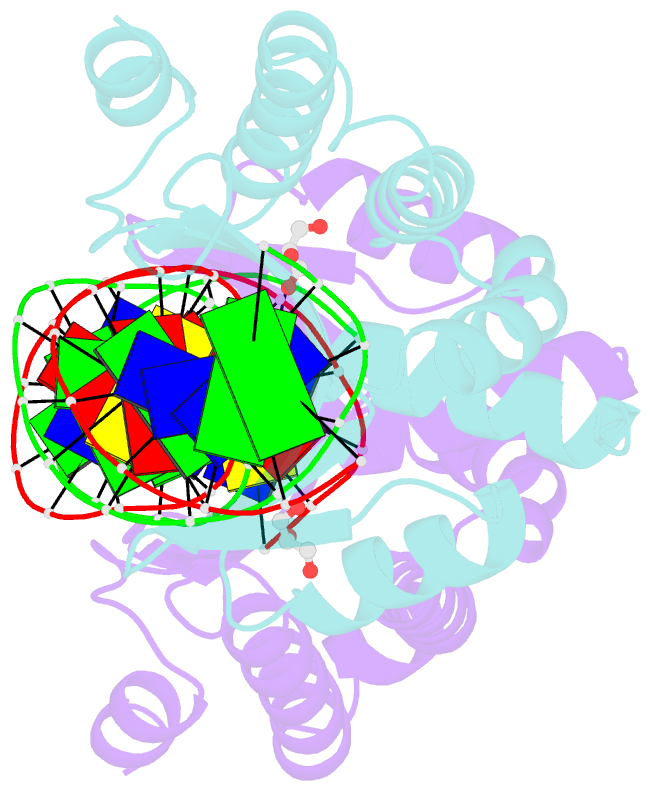
Summary information and primary citation
- PDB-id
- 4aqu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.3 Å)
- Summary
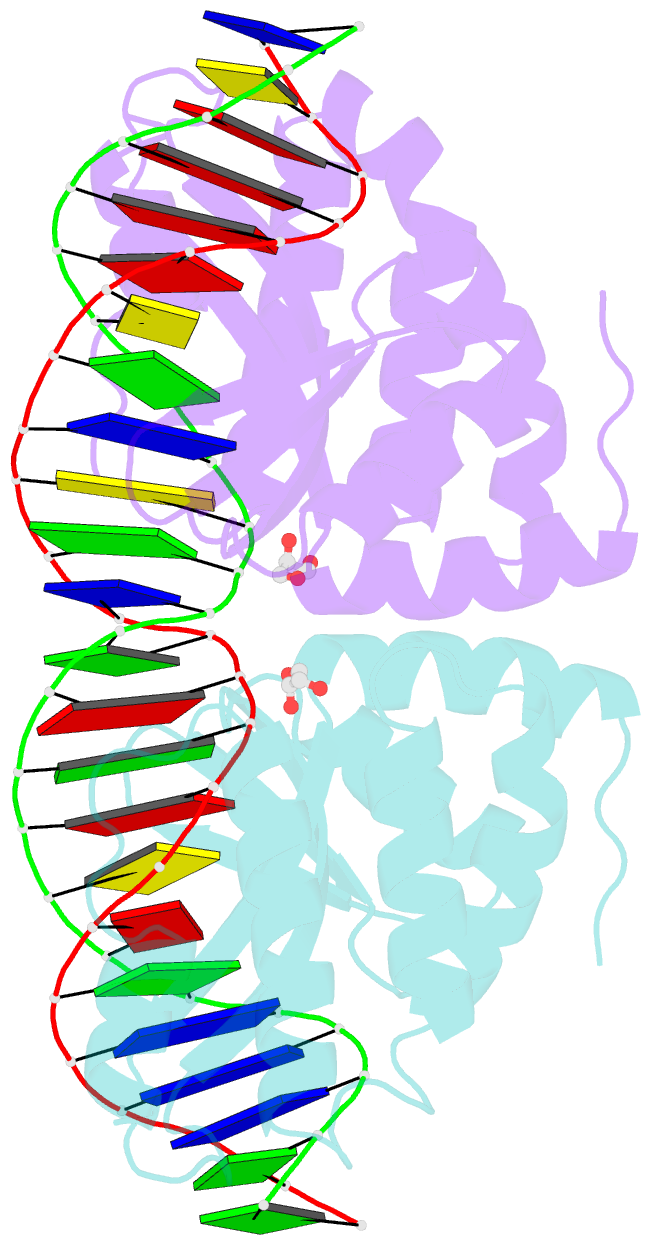
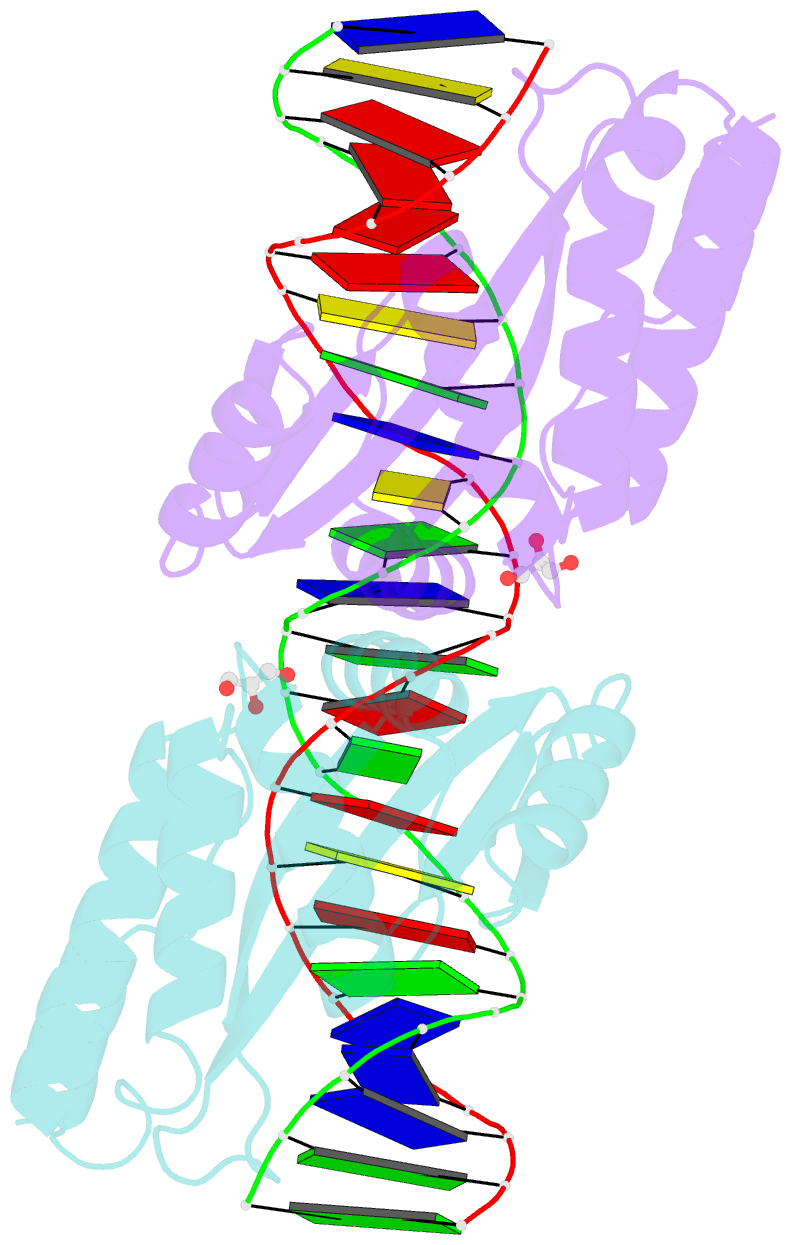
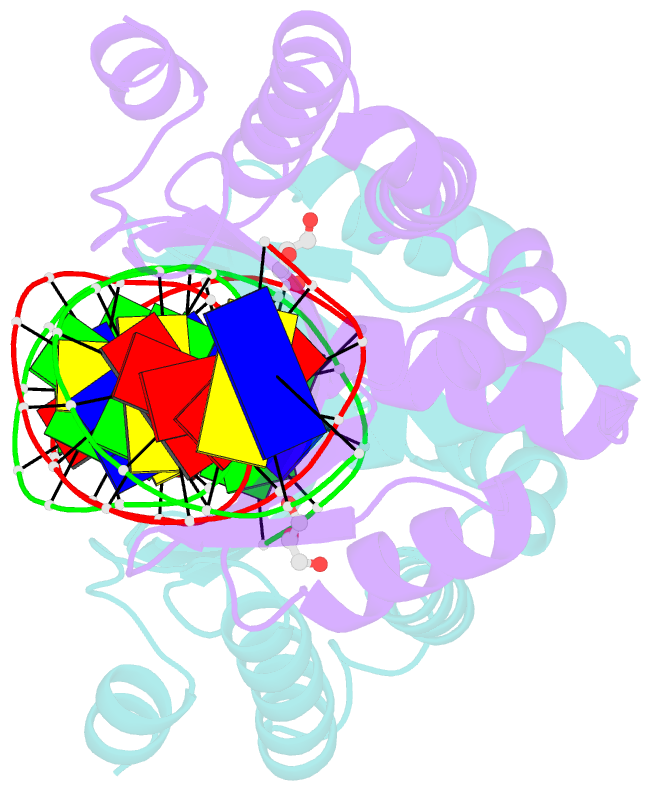
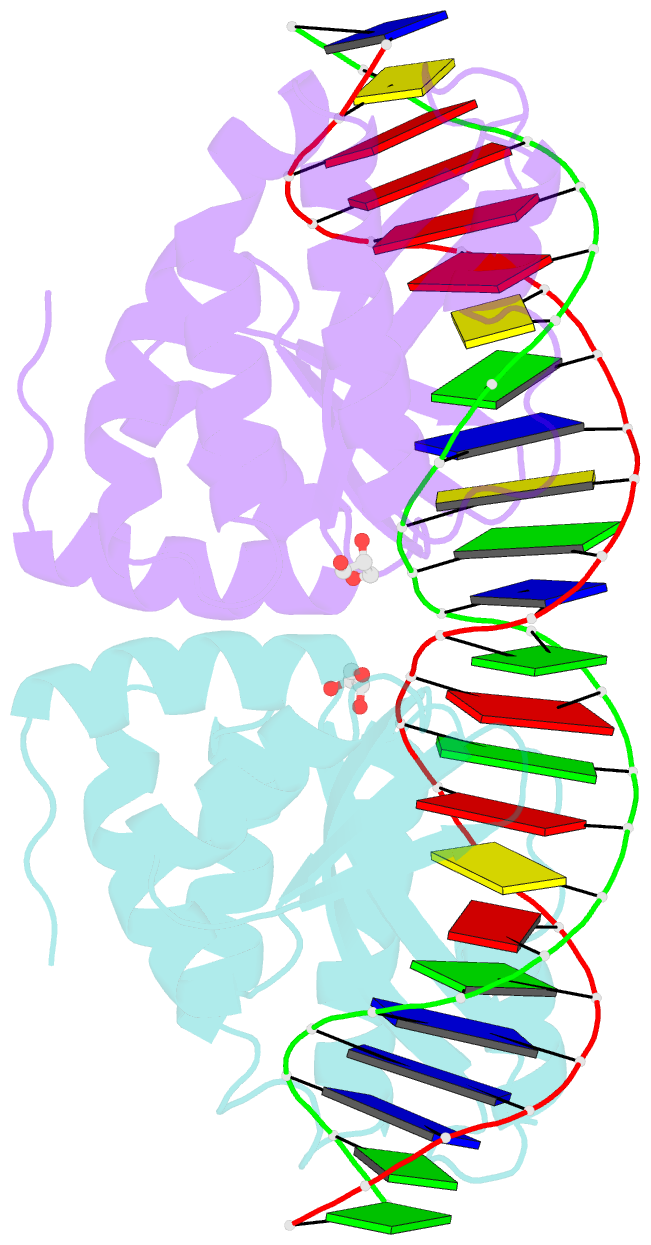
- Crystal structure of i-crei complexed with its target methylated at position plus 2 (in the b strand) in the presence of calcium
- Reference
- Valton J, Daboussi F, Leduc S, Molina R, Redondo P, Macmaster R, Montoya G, Duchateau P (2012): "5'-Cytosine-Phosphoguanine (Cpg) Methylation Impacts the Activity of Natural and Engineered Meganucleases." J.Biol.Chem., 287, 30139. doi: 10.1074/JBC.M112.379966.
- Abstract
- In this study, we asked whether CpG methylation could influence the DNA binding affinity and activity of meganucleases used for genome engineering applications. A combination of biochemical and structural approaches enabled us to demonstrate that CpG methylation decreases I-CreI DNA binding affinity and inhibits its endonuclease activity in vitro. This inhibition depends on the position of the methylated cytosine within the DNA target and was almost total when it is located inside the central tetrabase. Crystal structures of I-CreI bound to methylated cognate target DNA suggested a molecular basis for such inhibition, although the precise mechanism still has to be specified. Finally, we demonstrated that the efficacy of engineered meganucleases can be diminished by CpG methylation of the targeted endogenous site, and we proposed a rational design of the meganuclease DNA binding domain to alleviate such an effect. We conclude that although activity and sequence specificity of engineered meganucleases are crucial parameters, target DNA epigenetic modifications need to be considered for successful gene editions.