Summary information and primary citation
- PDB-id
- 4arc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.0 Å)
- Summary
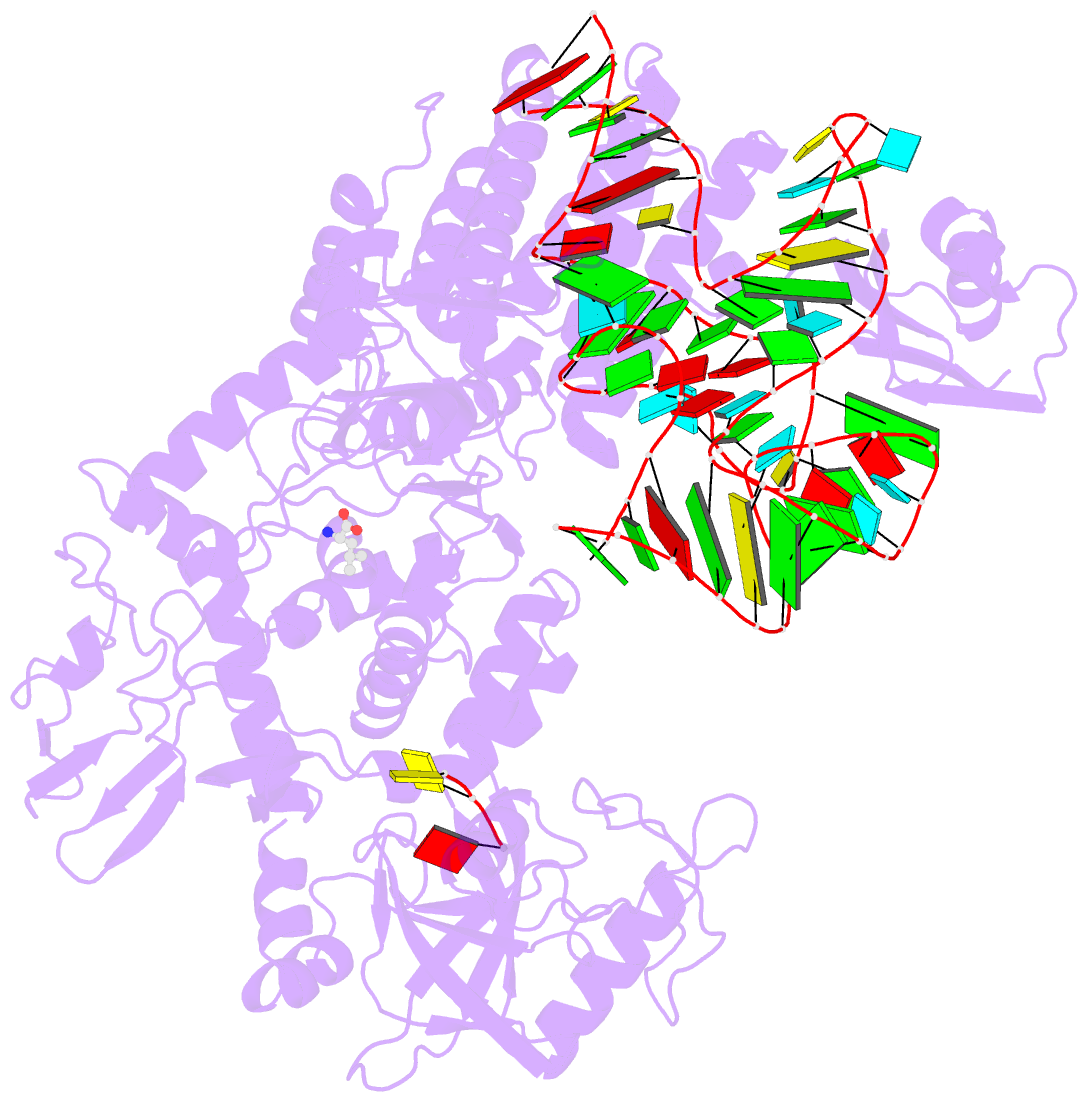
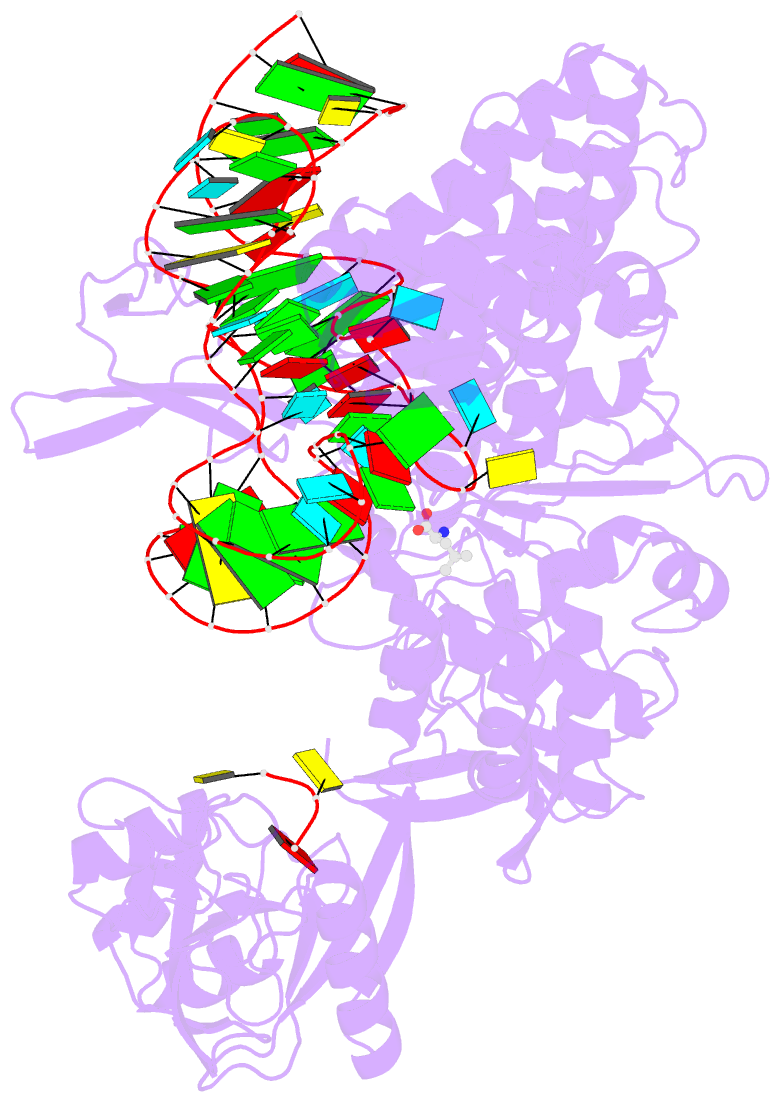
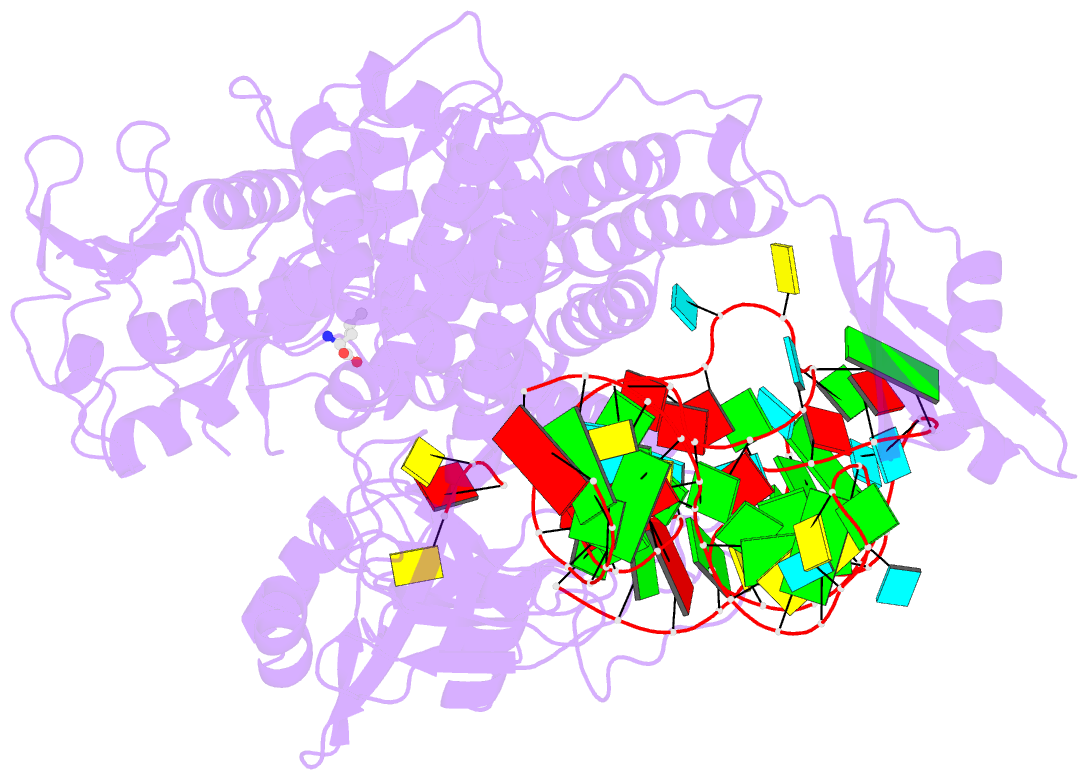
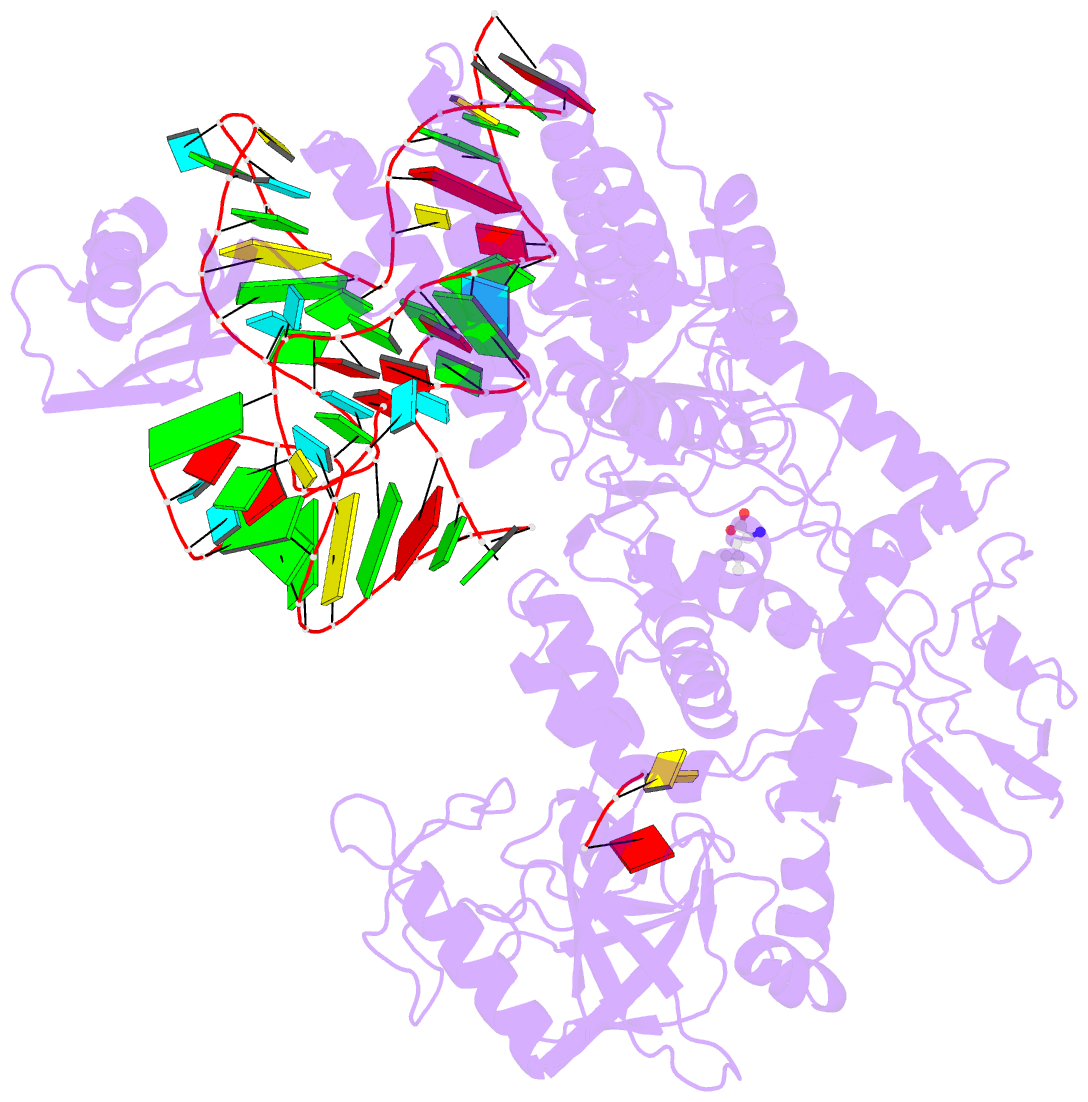
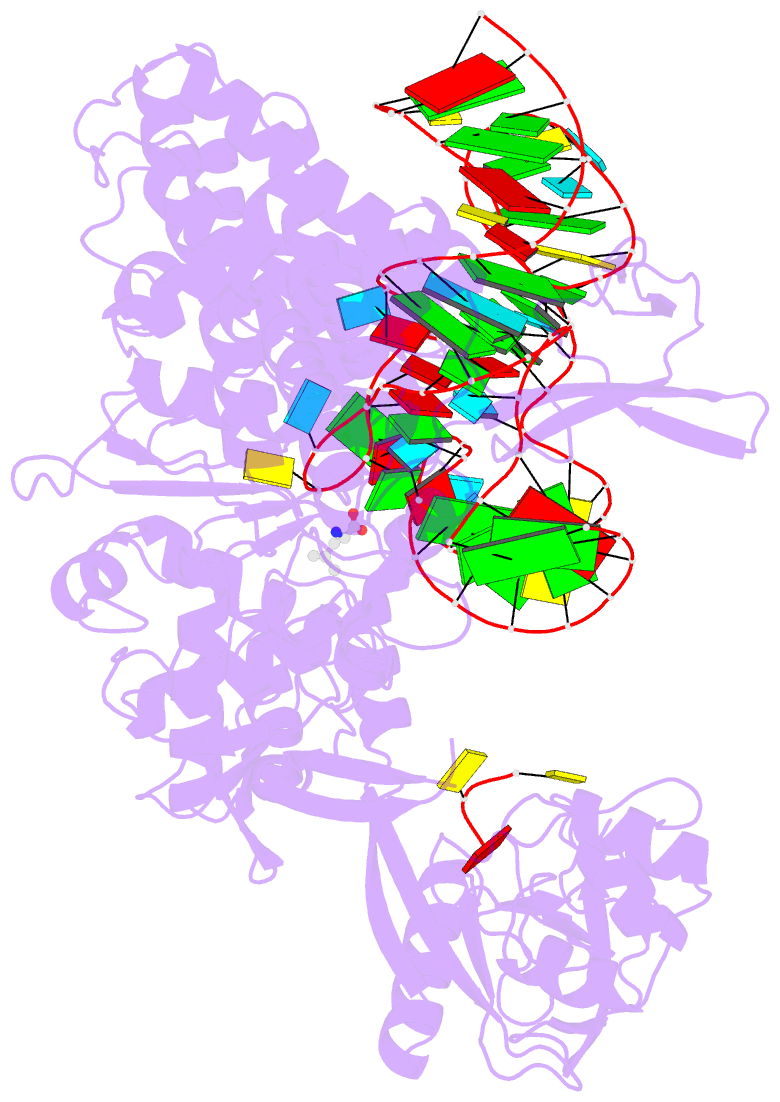
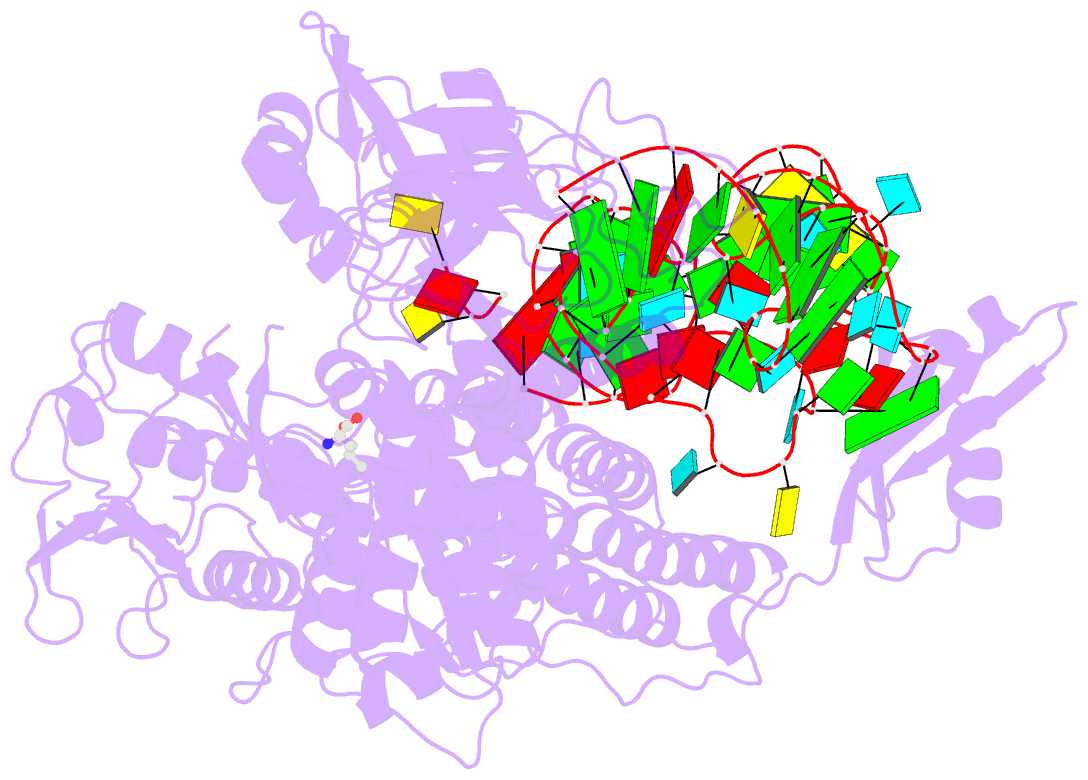
- Ternary complex of e. coli leucyl-trna synthetase, trna(leu) and leucine in the editing conformation
- Reference
- Palencia A, Crepin T, Vu MT, Lincecum Jr TL, Martinis SA, Cusack S (2012): "Structural Dynamics of the Aminoacylation and Proofreading Functional Cycle of Bacterial Leucyl-tRNA Synthetase." Nat.Struct.Mol.Biol., 19, 677. doi: 10.1038/NSMB.2317.
- Abstract
- Leucyl-tRNA synthetase (LeuRS) produces error-free leucyl-tRNA(Leu) by coordinating translocation of the 3' end of (mis-)charged tRNAs from its synthetic site to a separate proofreading site for editing. Here we report cocrystal structures of the Escherichia coli LeuRS-tRNA(Leu) complex in the aminoacylation or editing conformations, showing that translocation involves correlated rotations of four flexibly linked LeuRS domains. This pivots the tRNA to guide its charged 3' end from the closed aminoacylation state to the editing site. The editing domain unexpectedly stabilizes the tRNA during aminoacylation, and a large rotation of the leucine-specific domain positions the conserved KMSKS loop to bind the 3' end of the tRNA, promoting catalysis. Our results give new insight into the structural dynamics of a molecular machine that is essential for accurate protein synthesis.