Summary information and primary citation
- PDB-id
- 4b20; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.75 Å)
- Summary
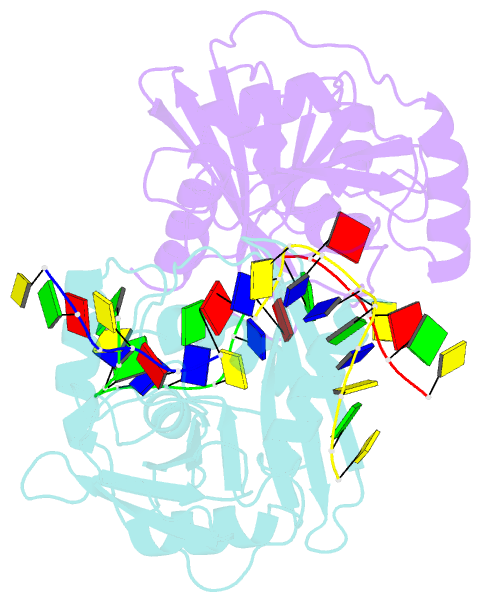
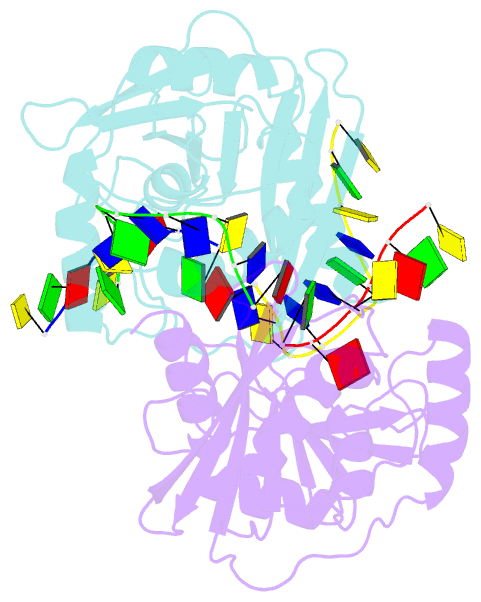
- Structural basis of DNA loop recognition by endonuclease v
- Reference
- Rosnes I, Rowe AD, Vik ES, Forstrom RJ, Alseth I, Bjoras M, Dalhus B (2013): "Structural Basis of DNA Loop Recognition by Endonuclease V." Structure, 21, 257. doi: 10.1016/J.STR.2012.12.007.
- Abstract
- The DNA repair enzyme endonuclease V (EndoV) recognizes and cleaves DNA at deaminated adenine lesions (hypoxanthine). In addition, EndoV cleaves DNA containing various helical distortions such as loops, hairpins, and flaps. To understand the molecular basis of EndoV's ability to recognize and incise DNA structures with helical distortions, we solved the crystal structure of Thermotoga maritima EndoV in complex with DNA containing a one-nucleotide loop. The structure shows that a strand-separating wedge is crucial for DNA loop recognition, with DNA strands separated precisely at the helical distortion. The additional nucleotide forming the loop rests on the surface of the wedge, while the normal adenine opposite the loop is flipped into a base recognition pocket. Our data show a different principle for DNA loop recognition and cleavage by EndoV, in which a coordinated action of a DNA-intercalating wedge and a base pocket accommodating a flipped normal base facilitate strand incision.