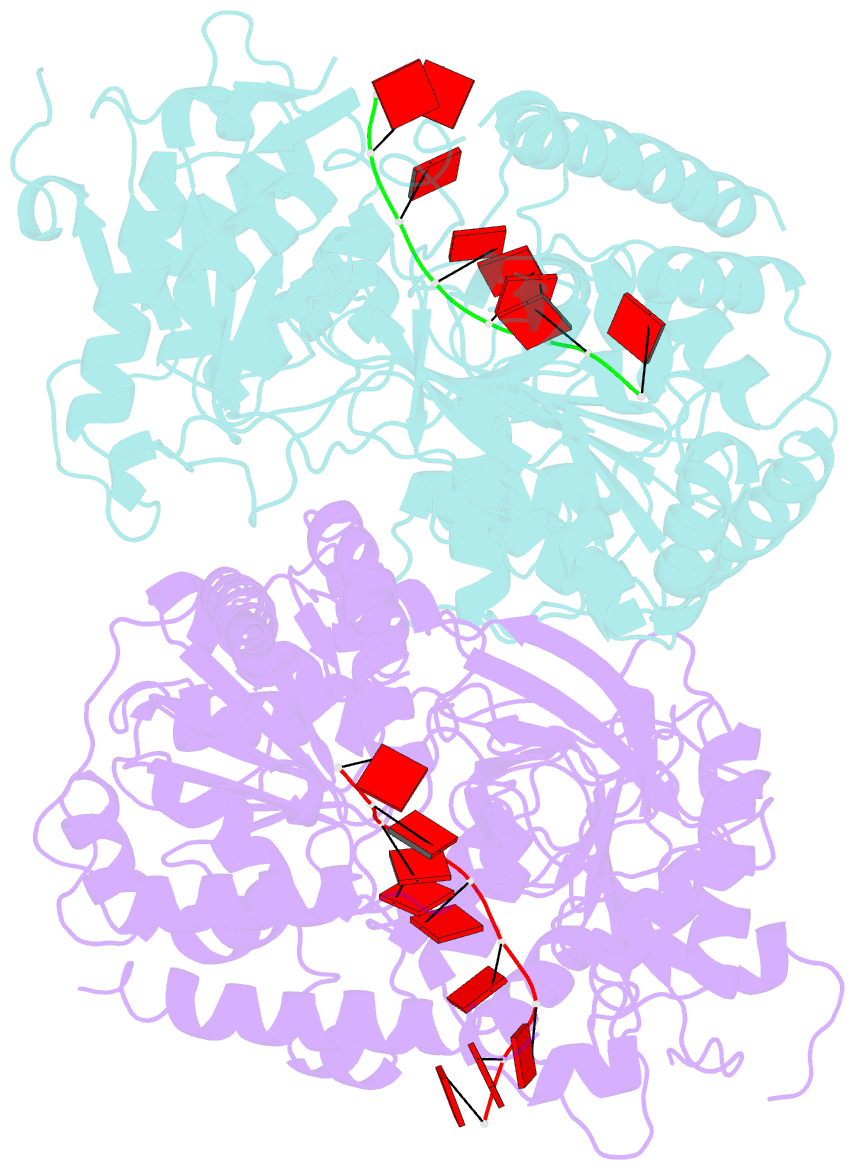
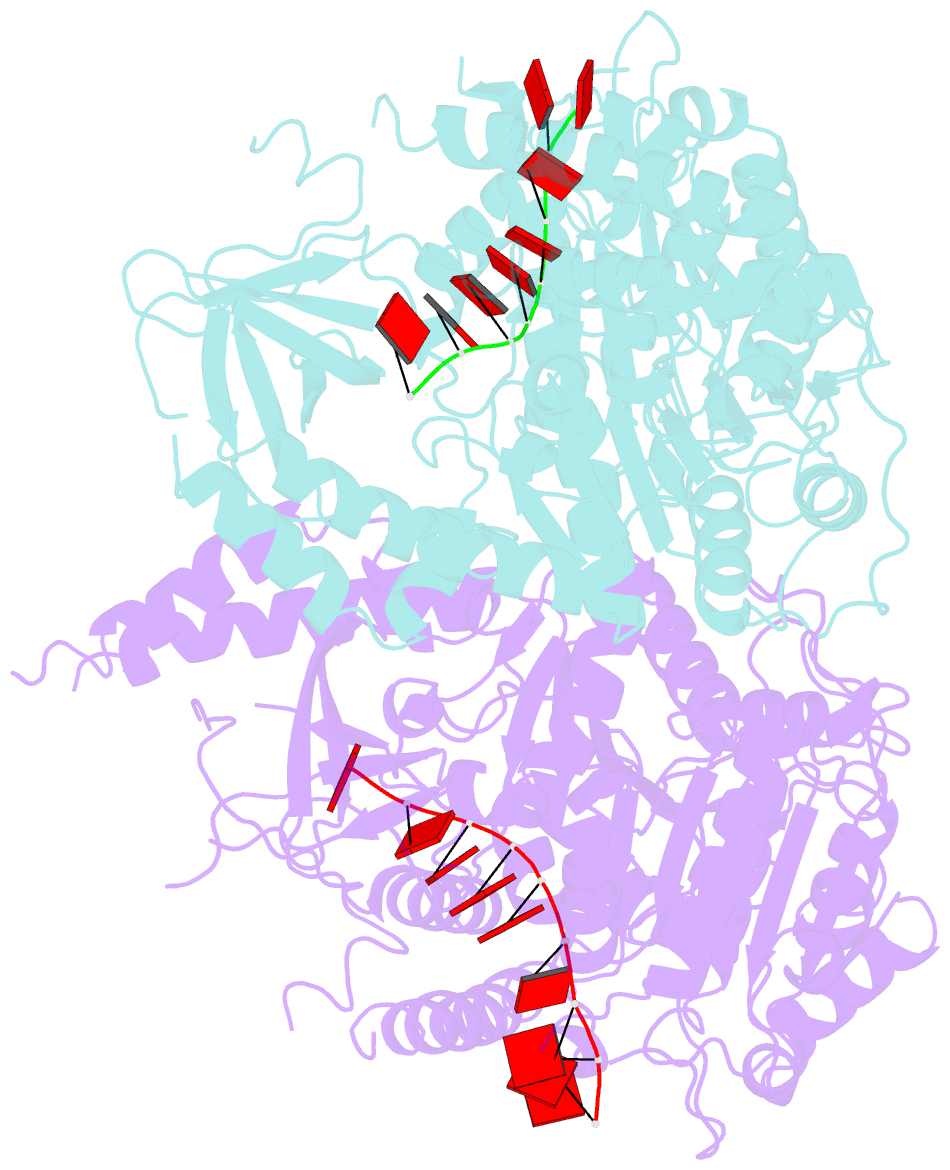
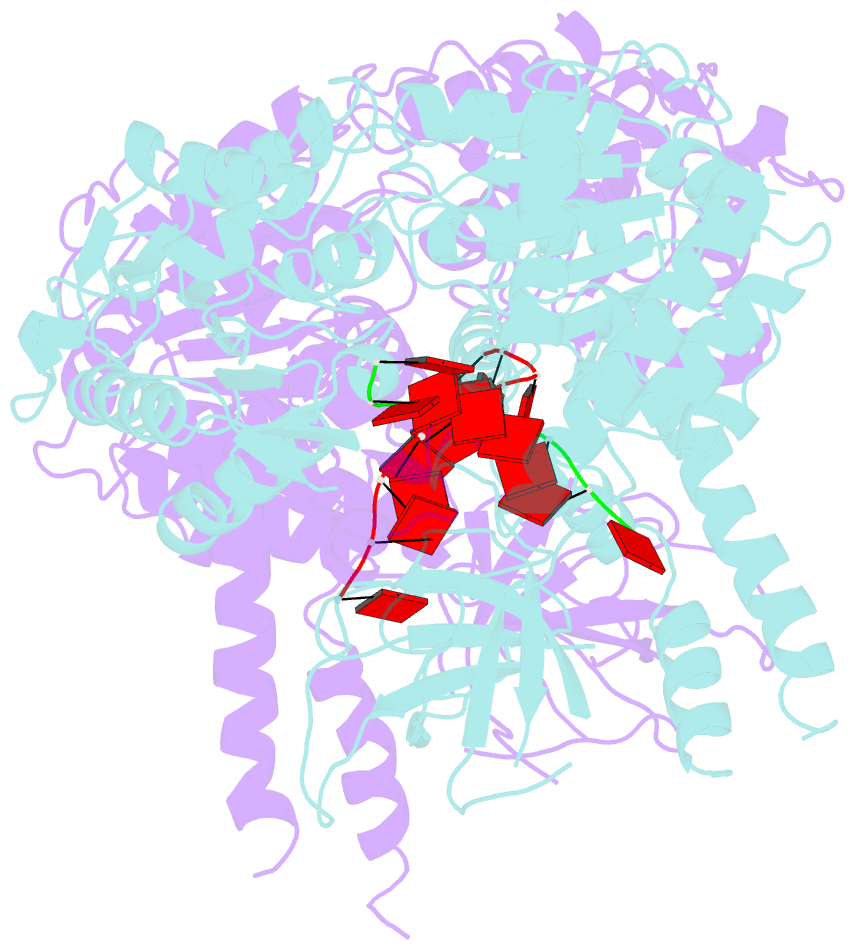
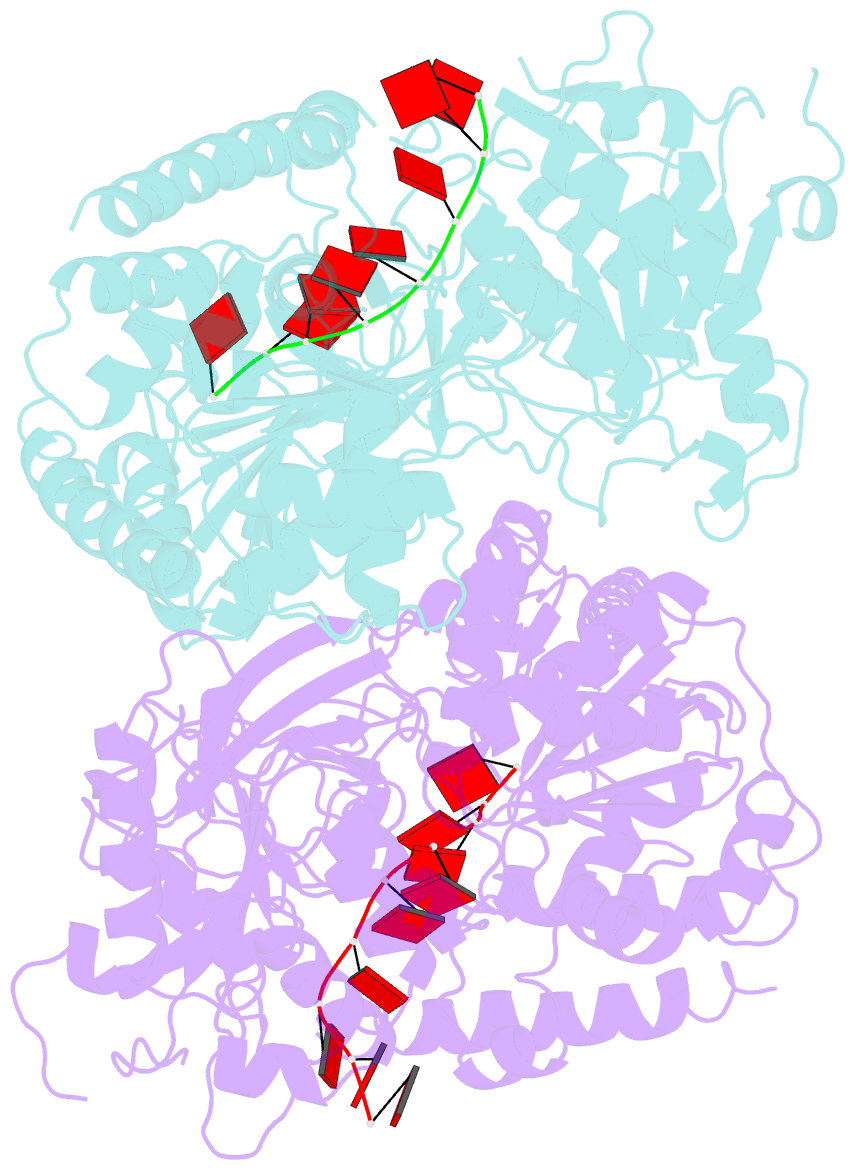
Summary information and primary citation
- PDB-id
- 4b3g; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.85 Å)
- Summary
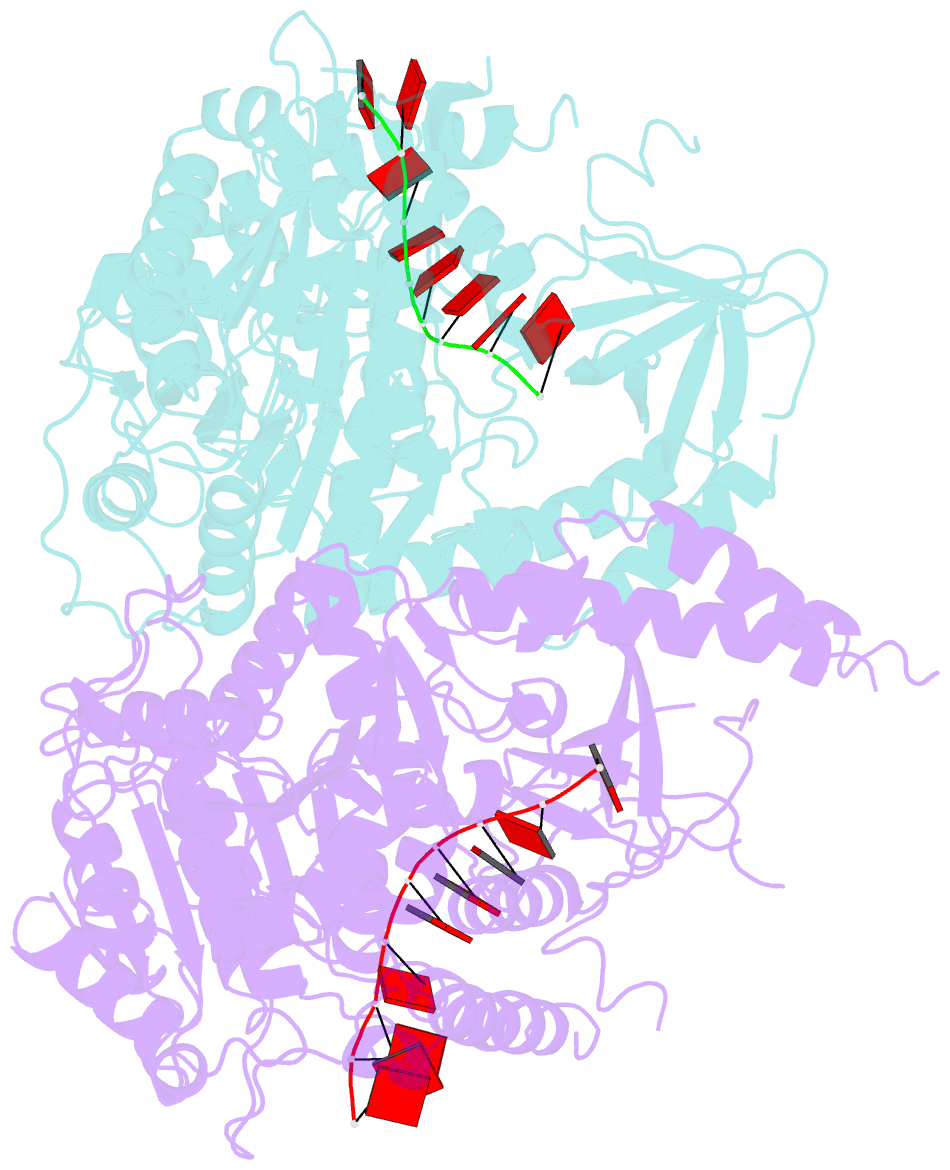
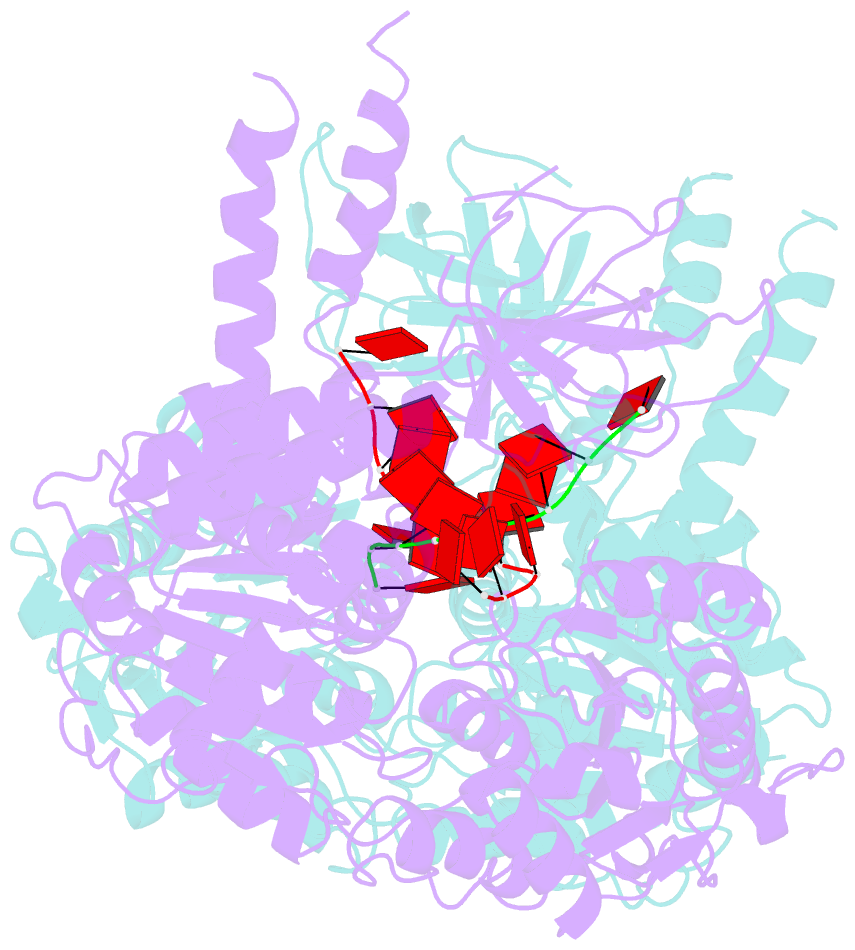
- Crystal structure of ighmbp2 helicase in complex with RNA
- Reference
- Lim SC, Bowler MW, Lai TF, Song H (2012): "The Ighmbp2 Helicase Structure Reveals the Molecular Basis for Disease-Causing Mutations in Dmsa1." Nucleic Acids Res., 40, 11009. doi: 10.1093/NAR/GKS792.
- Abstract
- Mutations in immunoglobulin µ-binding protein 2 (Ighmbp2) cause distal spinal muscular atrophy type 1 (DSMA1), an autosomal recessive disease that is clinically characterized by distal limb weakness and respiratory distress. However, despite extensive studies, the mechanism of disease-causing mutations remains elusive. Here we report the crystal structures of the Ighmbp2 helicase core with and without bound RNA. The structures show that the overall fold of Ighmbp2 is very similar to that of Upf1, a key helicase involved in nonsense-mediated mRNA decay. Similar to Upf1, domains 1B and 1C of Ighmbp2 undergo large conformational changes in response to RNA binding, rotating 30° and 10°, respectively. The RNA binding and ATPase activities of Ighmbp2 are further enhanced by the R3H domain, located just downstream of the helicase core. Mapping of the pathogenic mutations of DSMA1 onto the helicase core structure provides a molecular basis for understanding the disease-causing consequences of Ighmbp2 mutations.