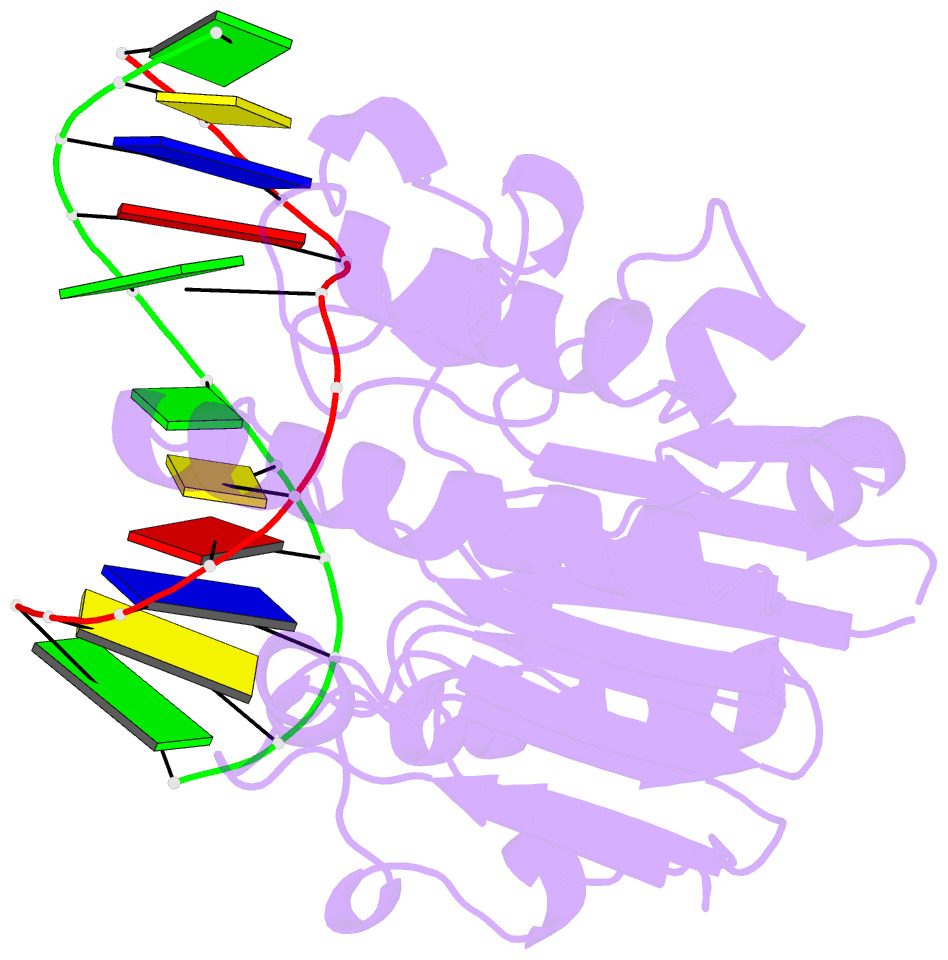
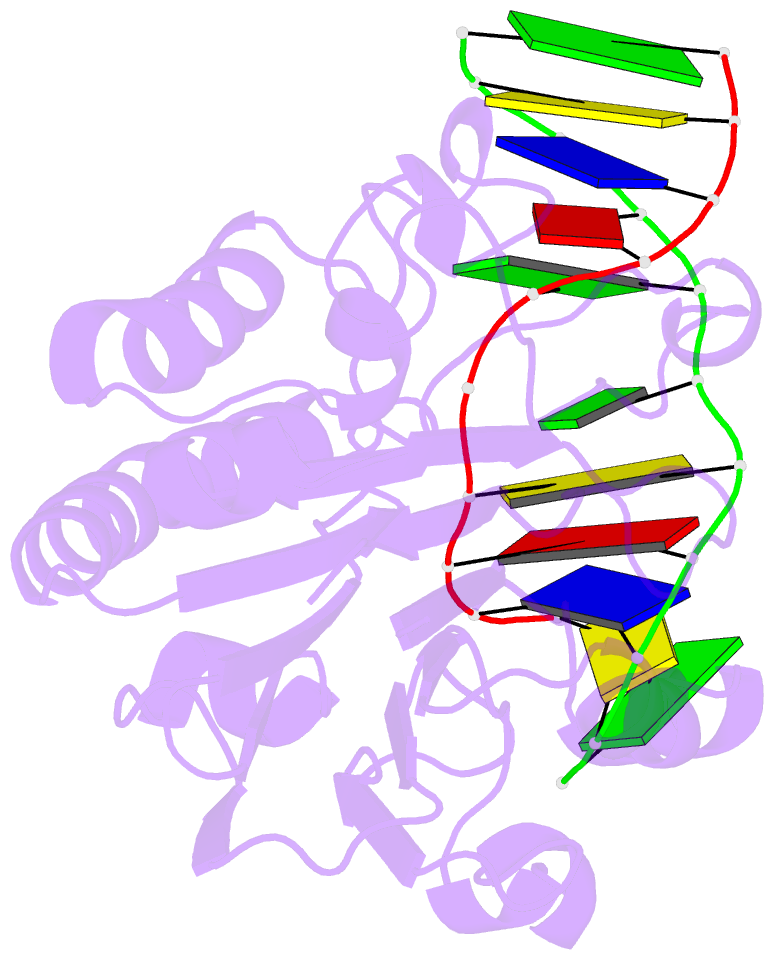
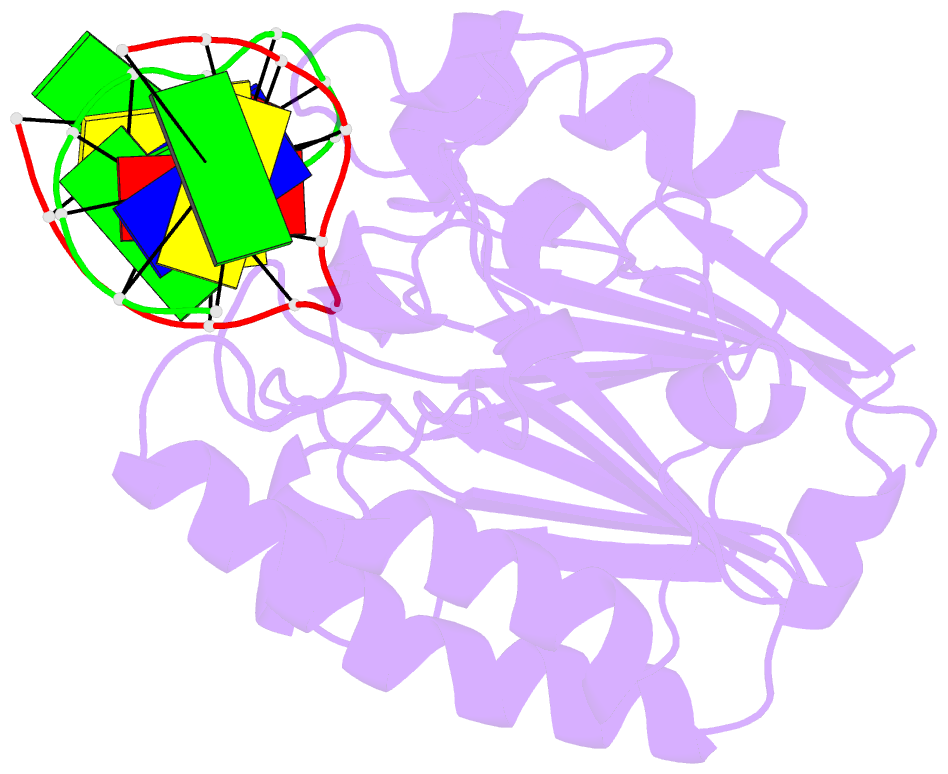
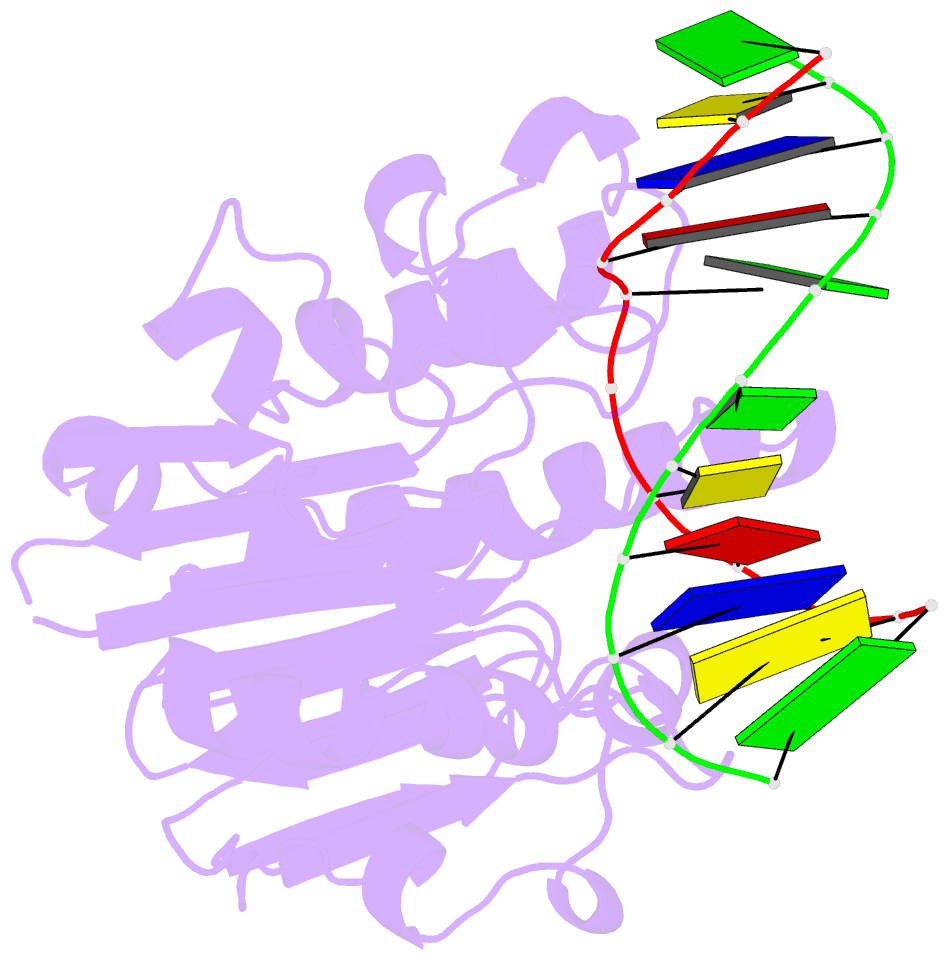
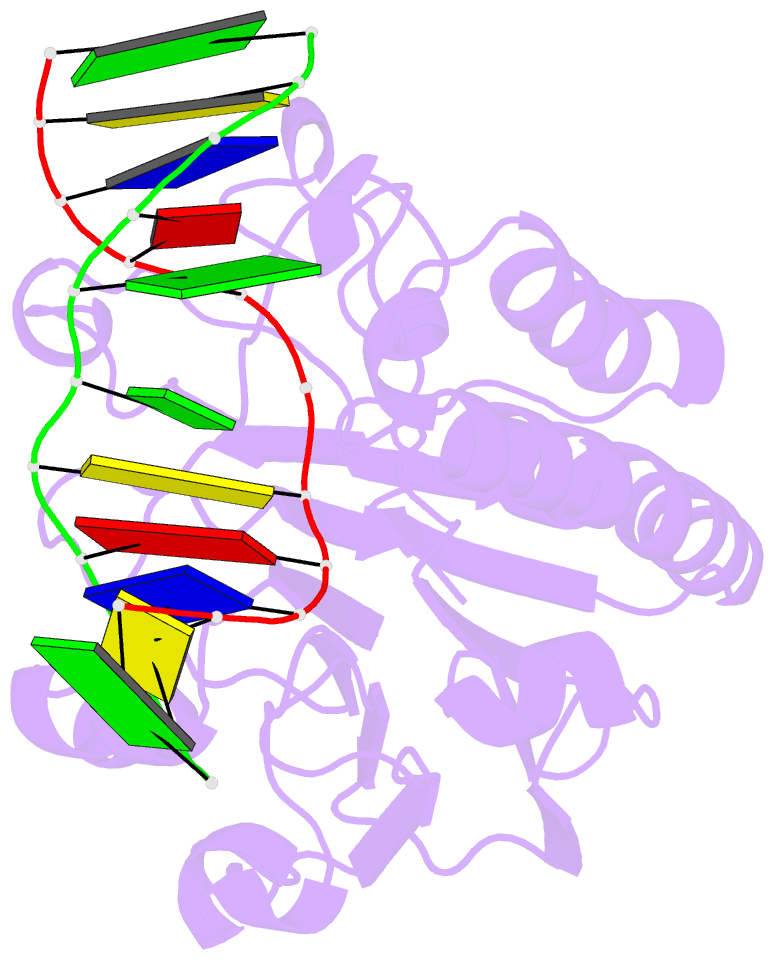
Summary information and primary citation
- PDB-id
- 4b5f; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.005 Å)
- Summary
- Substrate bound neisseria ap endonuclease in absence of metal ions (crystal form 1)
- Reference
- Lu D, Silhan J, MacDonald JT, Carpenter EP, Jensen K, Tang CM, Baldwin GS, Freemont PS (2012): "Structural basis for the recognition and cleavage of abasic DNA in Neisseria meningitidis." Proc. Natl. Acad. Sci. U.S.A., 109, 16852-16857. doi: 10.1073/pnas.1206563109.
- Abstract
- Base excision repair (BER) is a highly conserved DNA repair pathway throughout all kingdoms from bacteria to humans. Whereas several enzymes are required to complete the multistep repair process of damaged bases, apurinic-apyrimidic (AP) endonucleases play an essential role in enabling the repair process by recognizing intermediary abasic sites cleaving the phosphodiester backbone 5' to the abasic site. Despite extensive study, there is no structure of a bacterial AP endonuclease bound to substrate DNA. Furthermore, the structural mechanism for AP-site cleavage is incomplete. Here we report a detailed structural and biochemical study of the AP endonuclease from Neisseria meningitidis that has allowed us to capture structural intermediates providing more complete snapshots of the catalytic mechanism. Our data reveal subtle differences in AP-site recognition and kinetics between the human and bacterial enzymes that may reflect different evolutionary pressures.