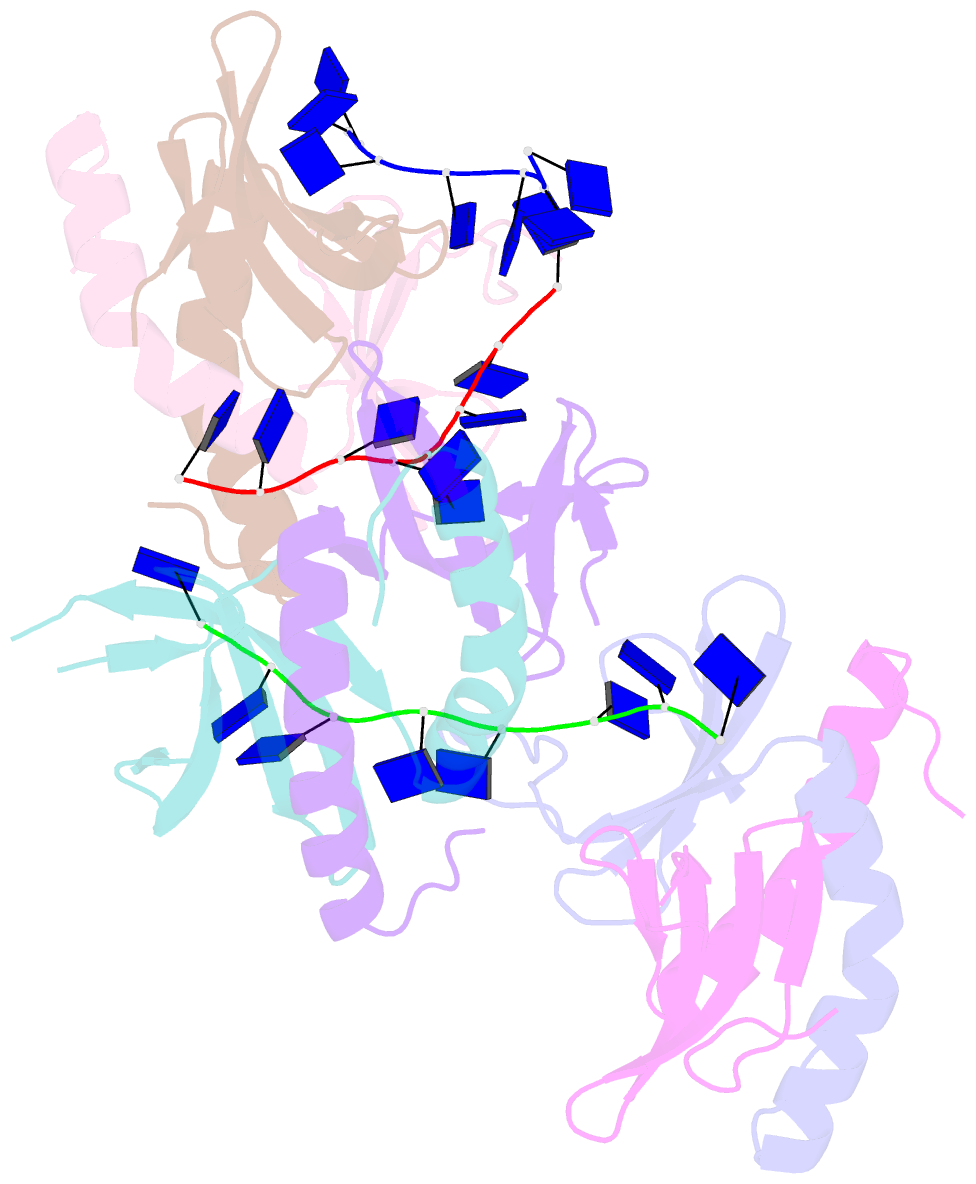
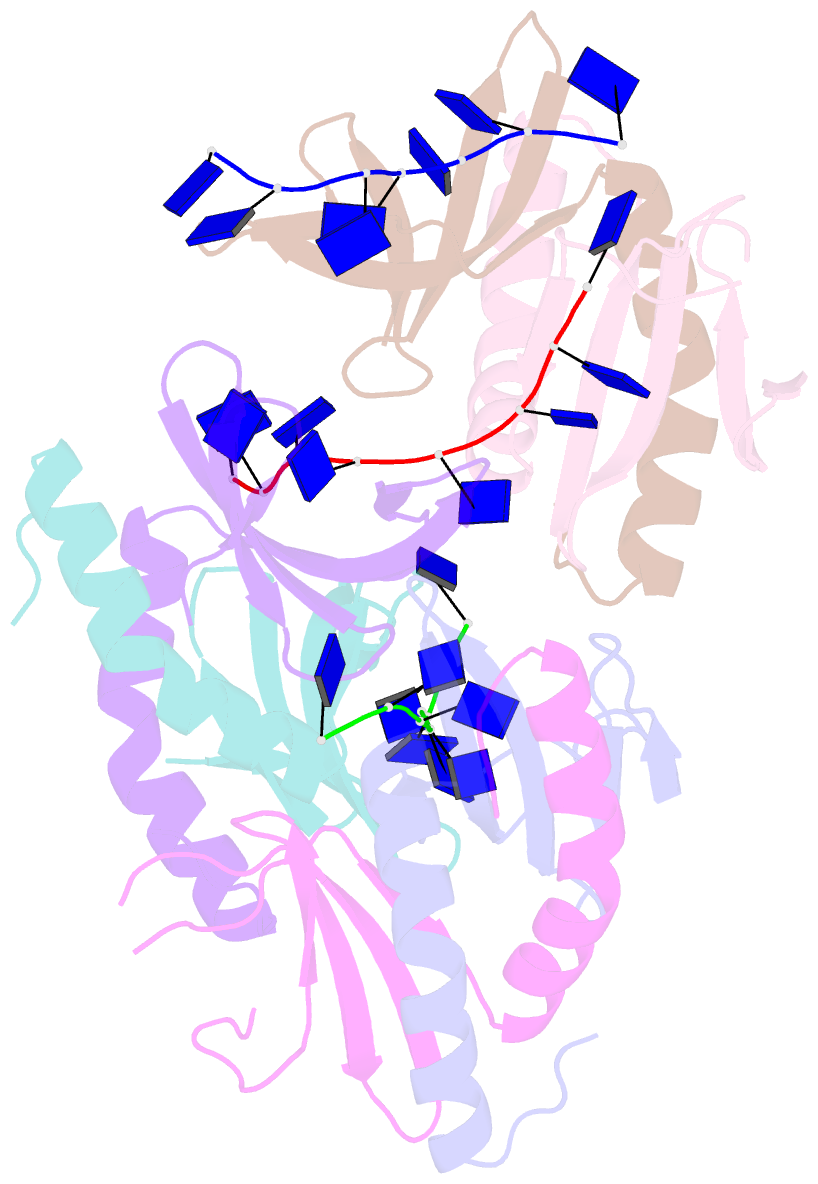
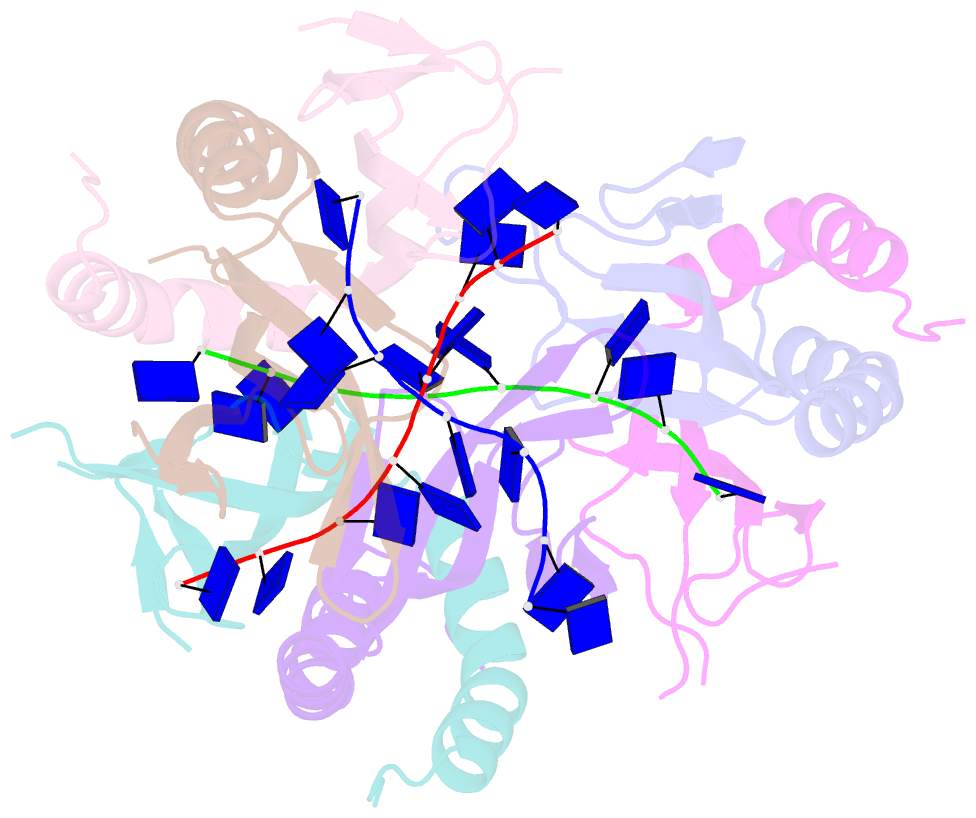
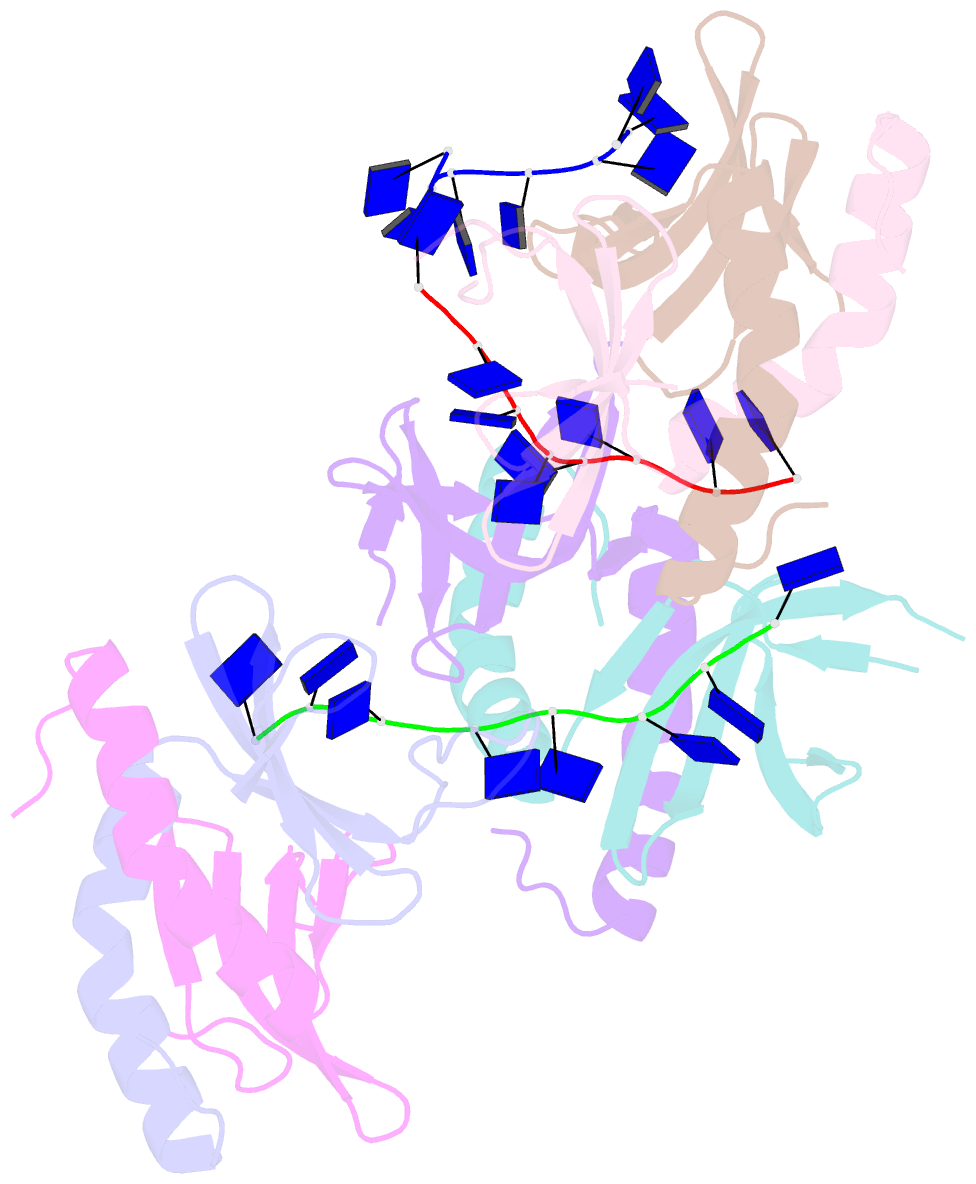
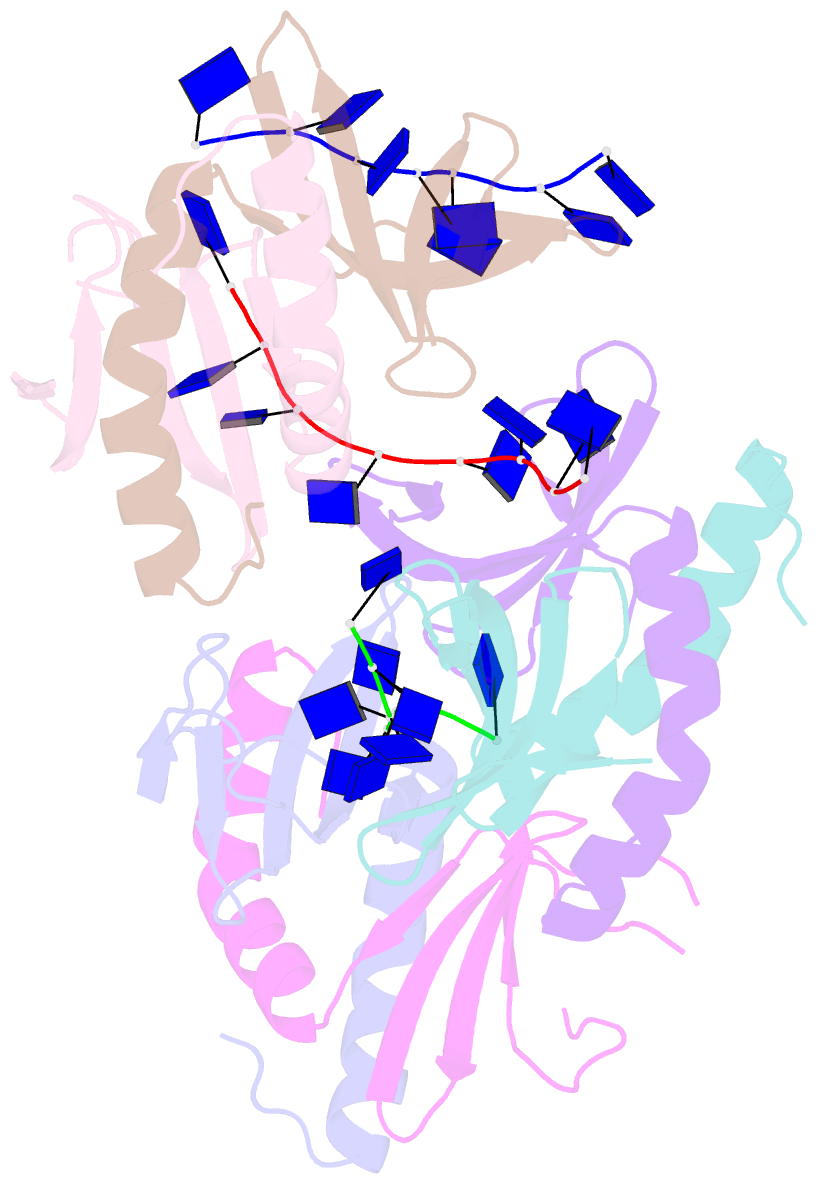
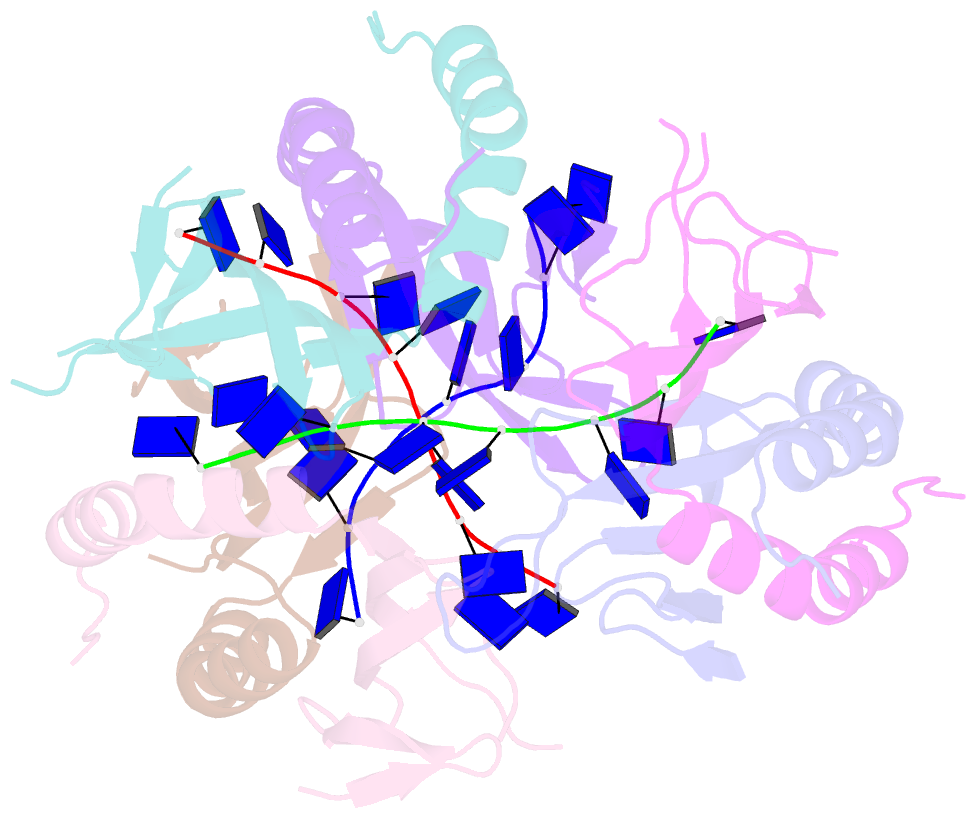
Summary information and primary citation
- PDB-id
- 4bhm; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.7 Å)
- Summary
- The crystal structure of mosub1-DNA complex reveals a novel DNA binding mode
- Reference
- Huang J, Zhao Y, Liu H, Huang D, Cheng X, Zhao W, Taylor IA, Liu J, Peng YL (2015): "Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4." Sci.Rep., 5, 8789. doi: 10.1038/SREP08789.
- Abstract
- PC4, a well-known general transcription cofactor, has multiple functions in transcription and DNA repair. Residue W89, is engaged in stacking interactions with DNA in PC4, but substituted by tyrosine in some PC4 orthologous proteins. In order to understand the consequences and reveal the molecular details of this substitution we have determined the crystal structures of the PC4 orthologue MoSub1 and a PC4 W89Y mutant in complex with DNA. In the structure of MoSub1-DNA complex, Y74 interacts directly with a single nucleotide of oligo DNA. By comparison, the equivalent residue, W89 in wild type PC4 interacts with two nucleotides and the base of the second nucleotide has distinct orientation relative to that of the first one. A hydrophobic patch around W89 that favours interaction with two nucleotides is not formed in the PC4 W89Y mutant. Therefore, the change of the surface hydrophobicity around residue 89 results in a difference between the modes of DNA interaction. These results indicate that the conserved Y74 in MoSub1 or W89 in PC4, are not only key residues in making specific interactions with DNA but also required to determine the DNA binding mode of PC4 proteins.