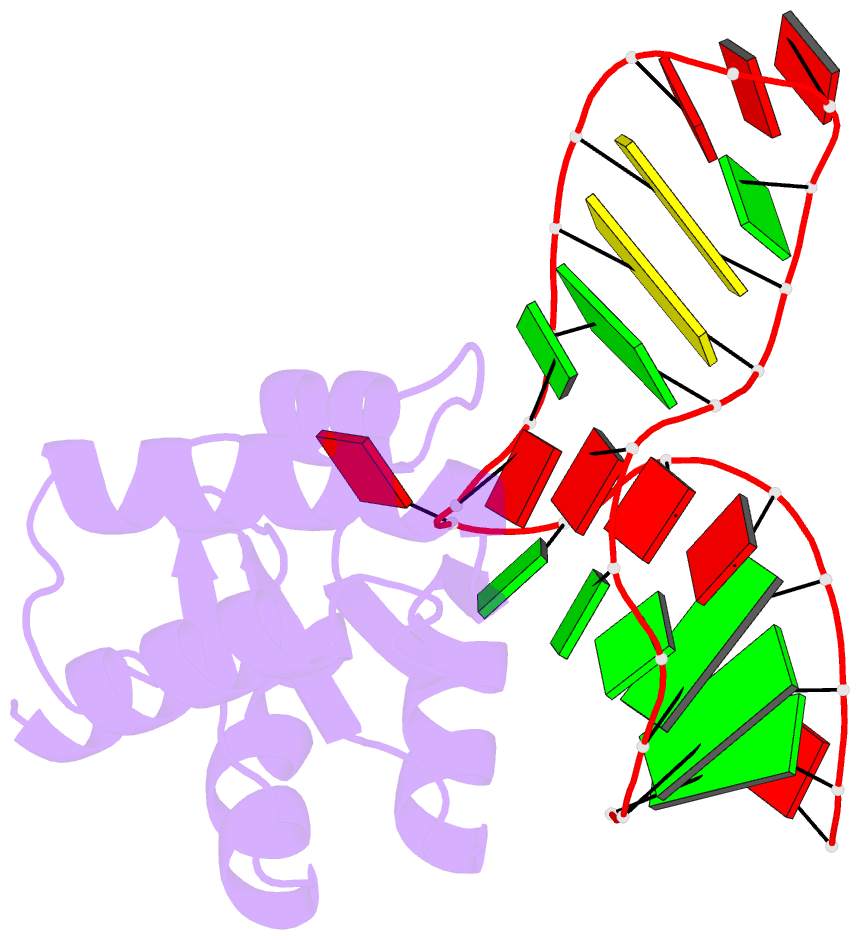
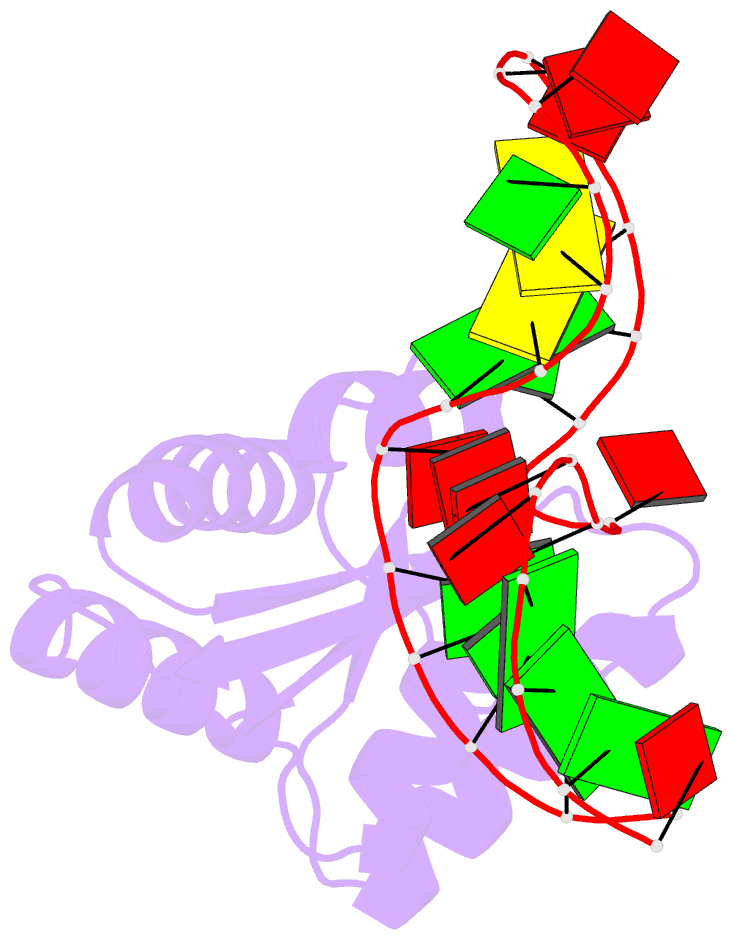
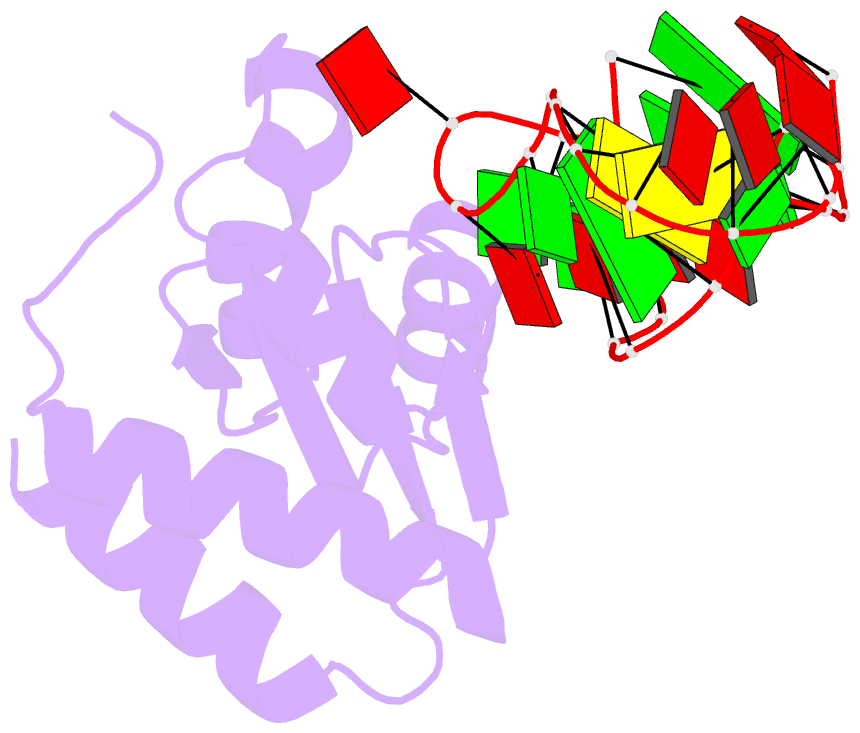
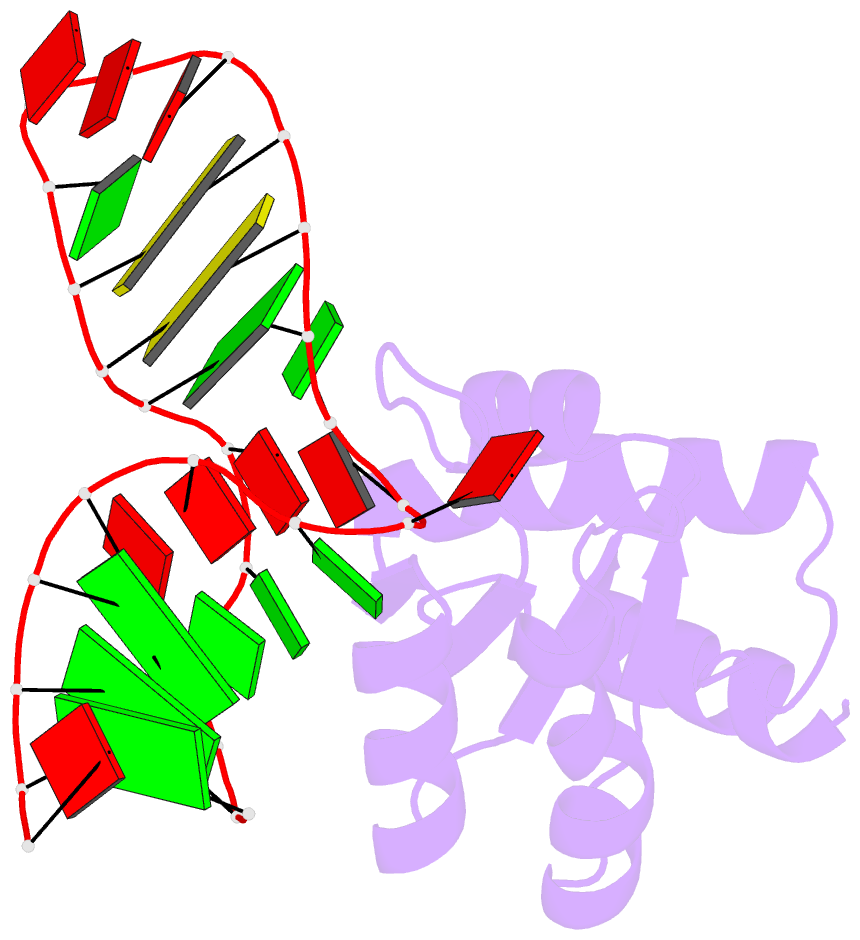
Summary information and primary citation
- PDB-id
- 4bw0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-RNA binding protein
- Method
- X-ray (2.33 Å)
- Summary
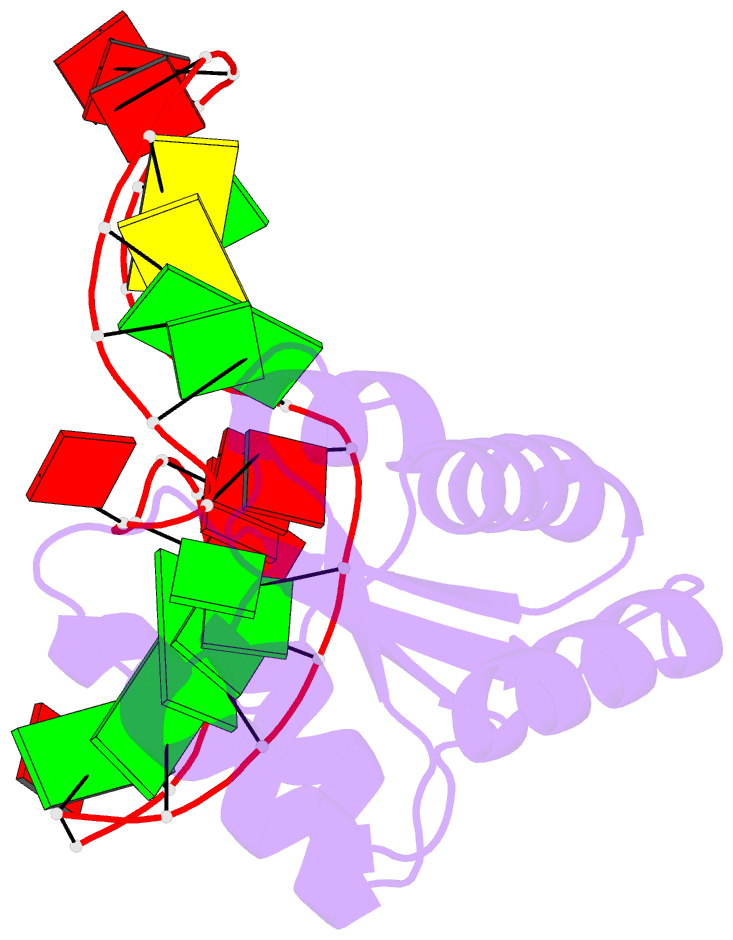
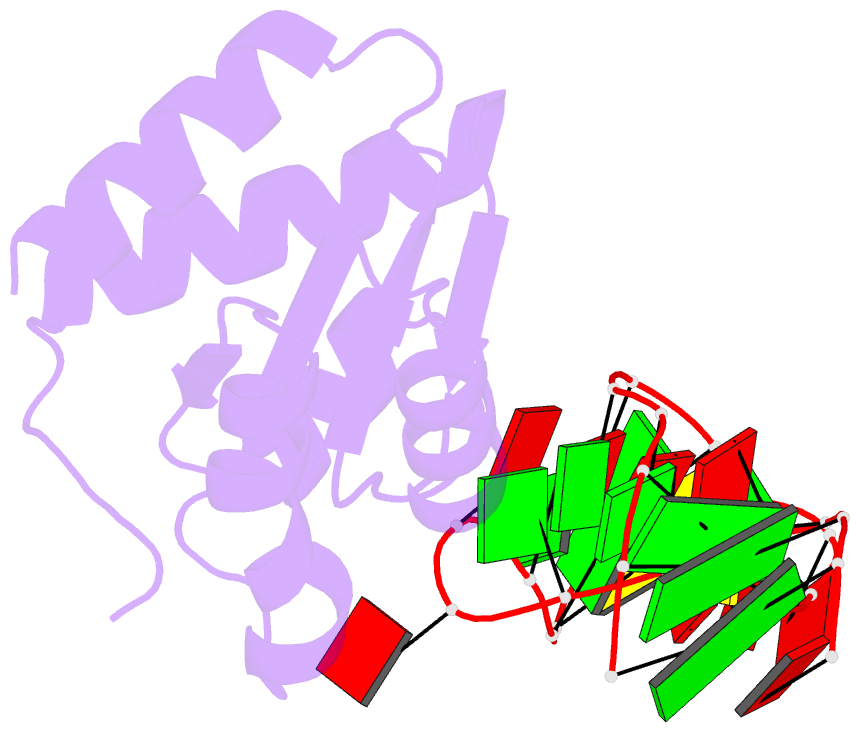
- The molecular recognition of kink turn structure by the l7ae class of proteins
- Reference
- Huang L, Lilley DMJ (2013): "The Molecular Recognition of Kink-Turn Structure by the L7Ae Class of Proteins." RNA, 19, 1703. doi: 10.1261/RNA.041517.113.
- Abstract
- L7Ae is a member of a protein family that binds kink-turns (k-turns) in many functional RNA species. We have solved the X-ray crystal structure of the near-consensus sequence Kt-7 of Haloarcula marismortui bound by Archaeoglobus fulgidus L7Ae at 2.3-Å resolution. We also present a structure of Kt-7 in the absence of bound protein at 2.2-Å resolution. As a result, we can describe a general mode of recognition of k-turn structure by the L7Ae family proteins. The protein makes interactions in the widened major groove on the outer face of the k-turn. Two regions of the protein are involved. One is an α-helix that enters the major groove of the NC helix, making both nonspecific backbone interactions and specific interactions with the guanine nucleobases of the conserved G • A pairs. A hydrophobic loop makes close contact with the L1 and L2 bases, and a glutamate side chain hydrogen bonds with L1. Taken together, these interactions are highly selective for the structure of the k-turn and suggest how conformational selection of the folded k-turn occurs.