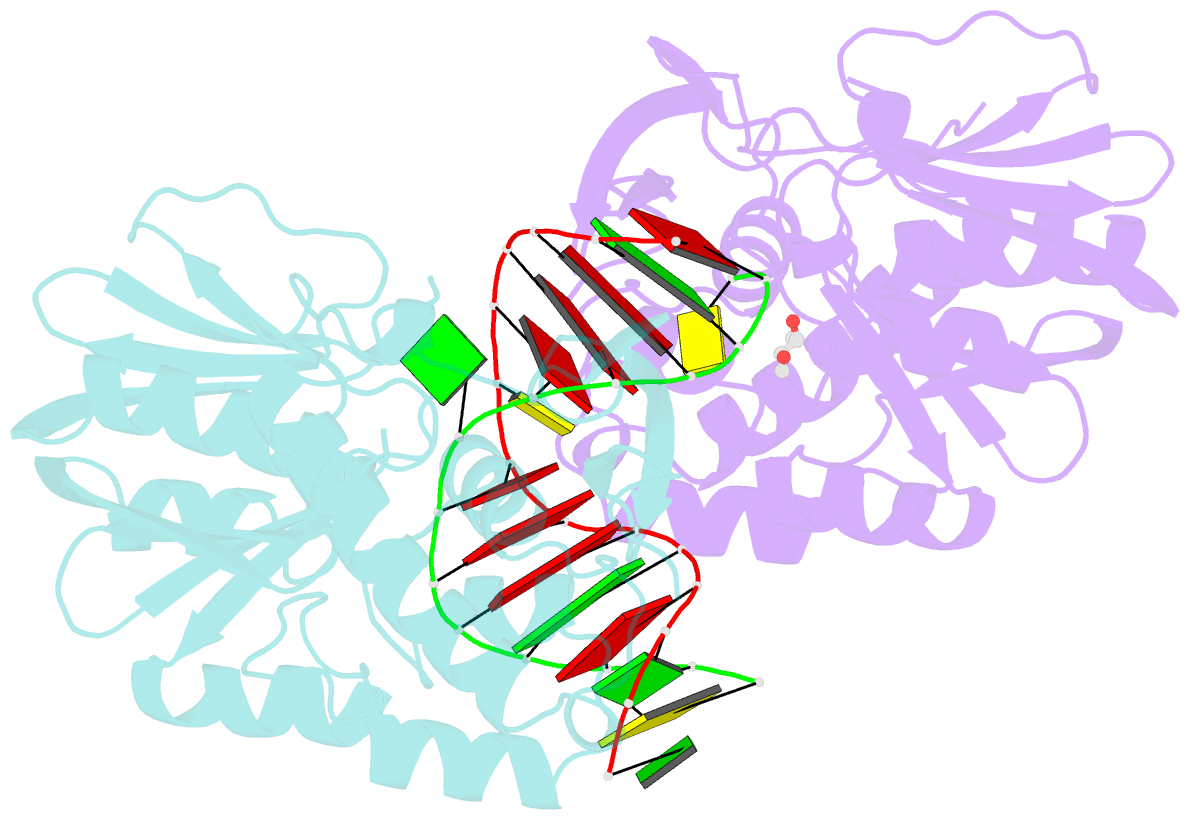
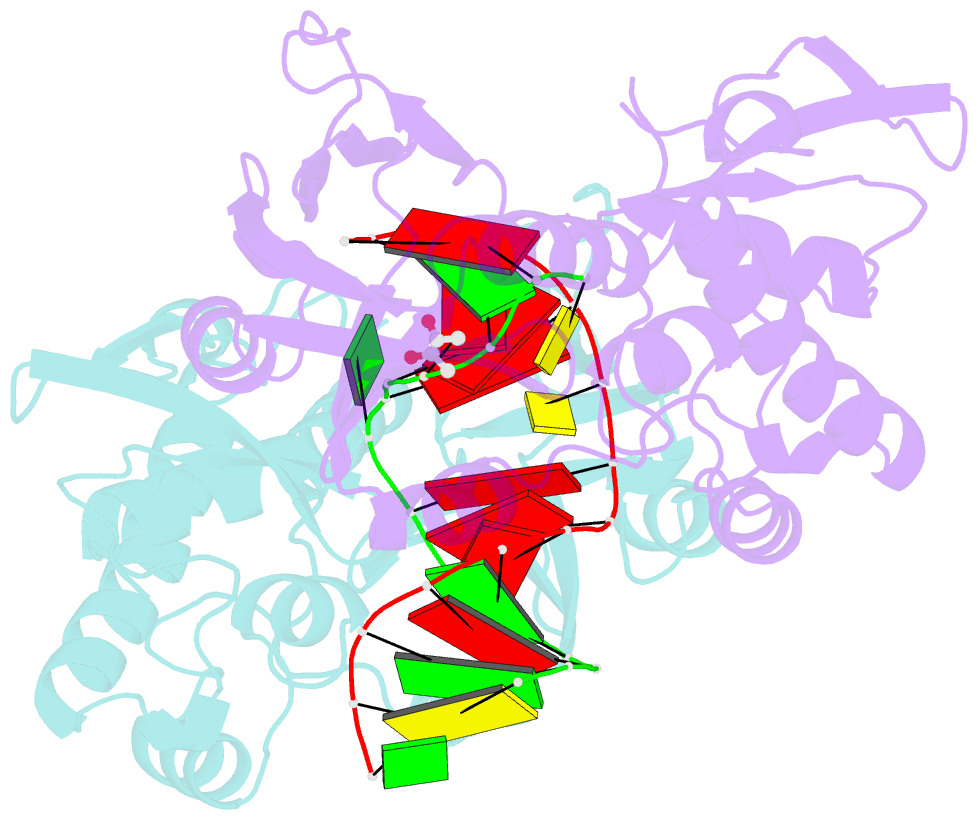
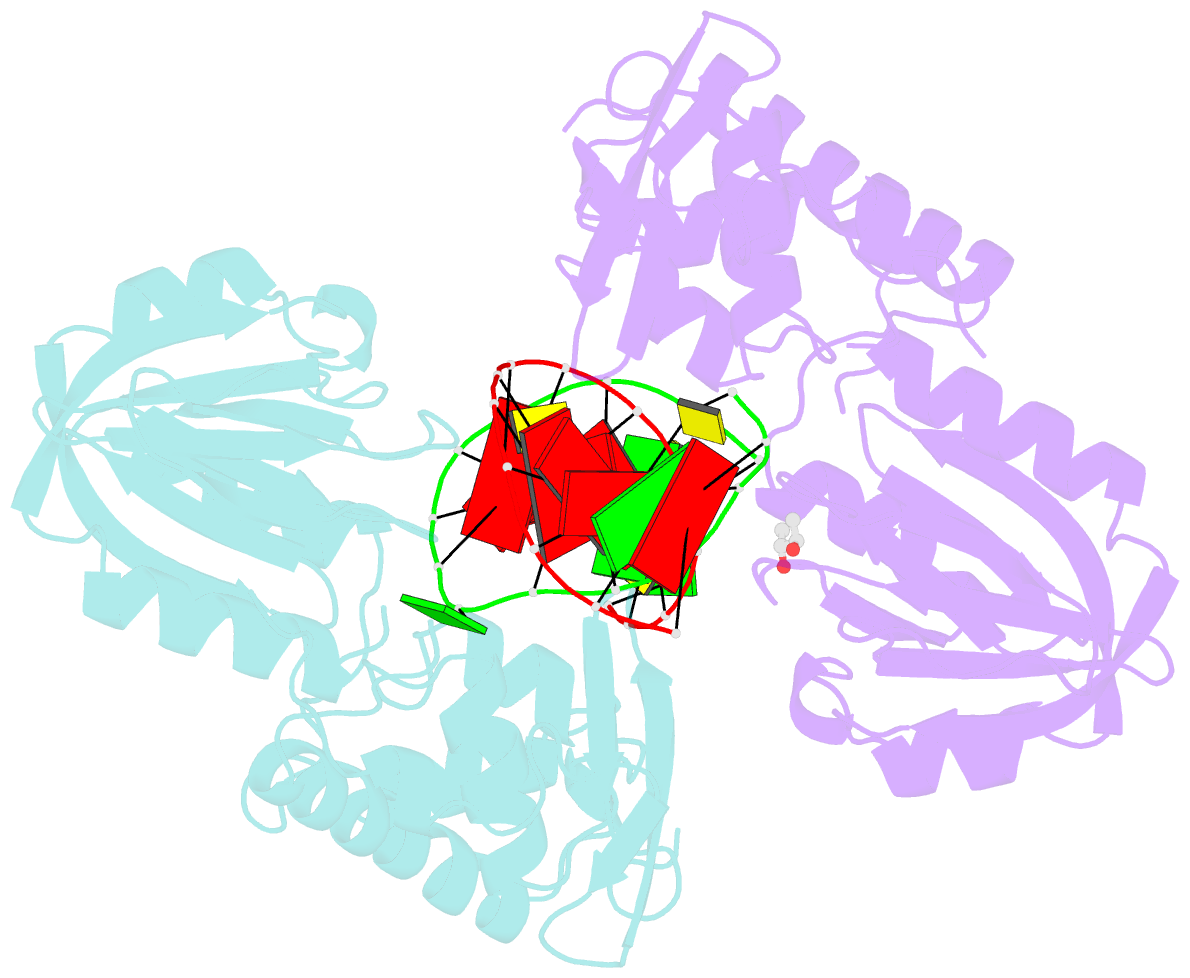
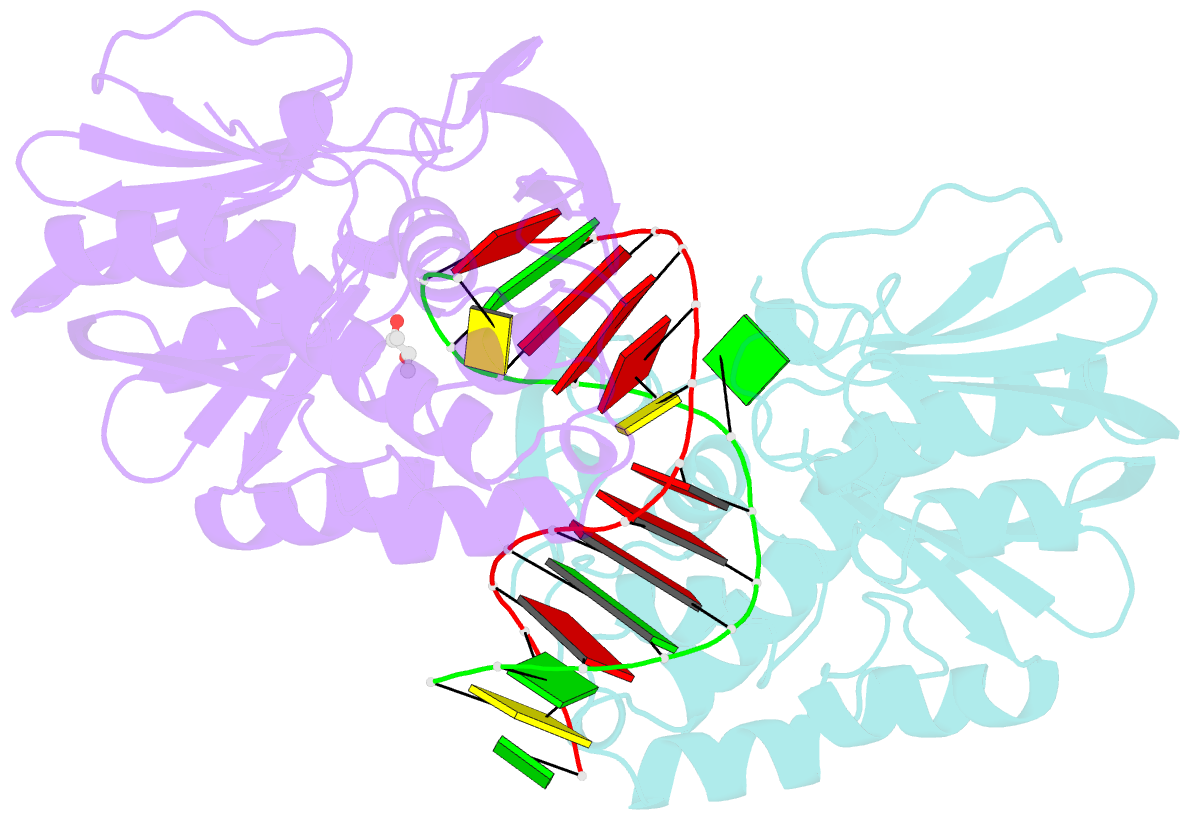
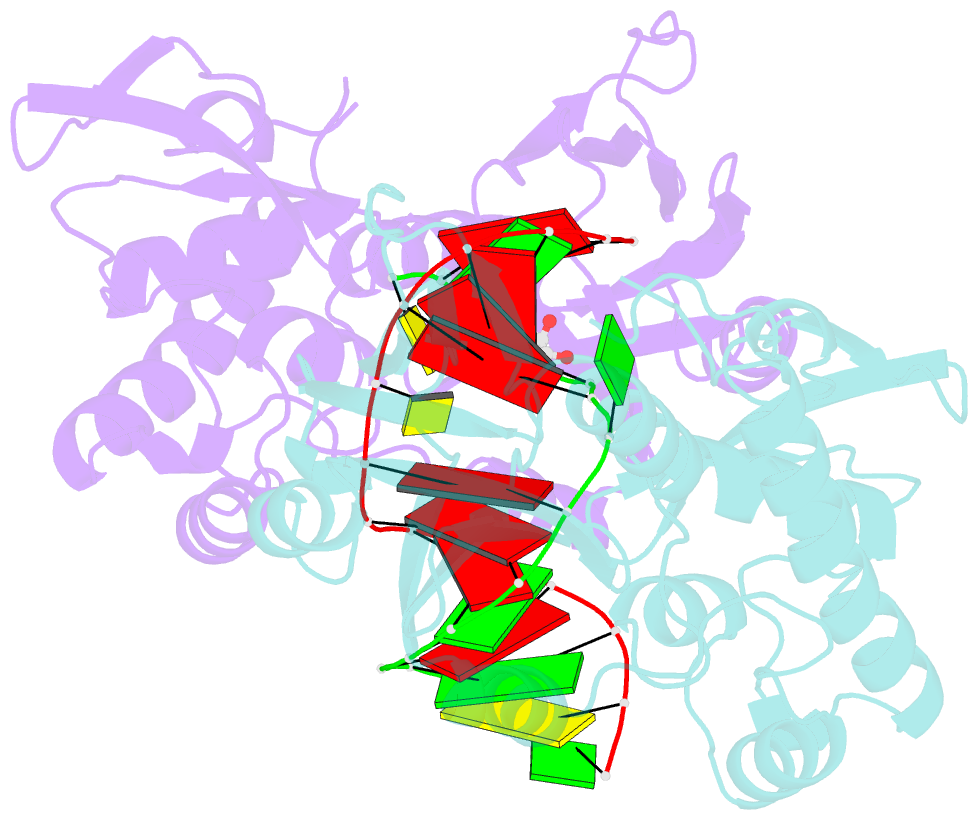
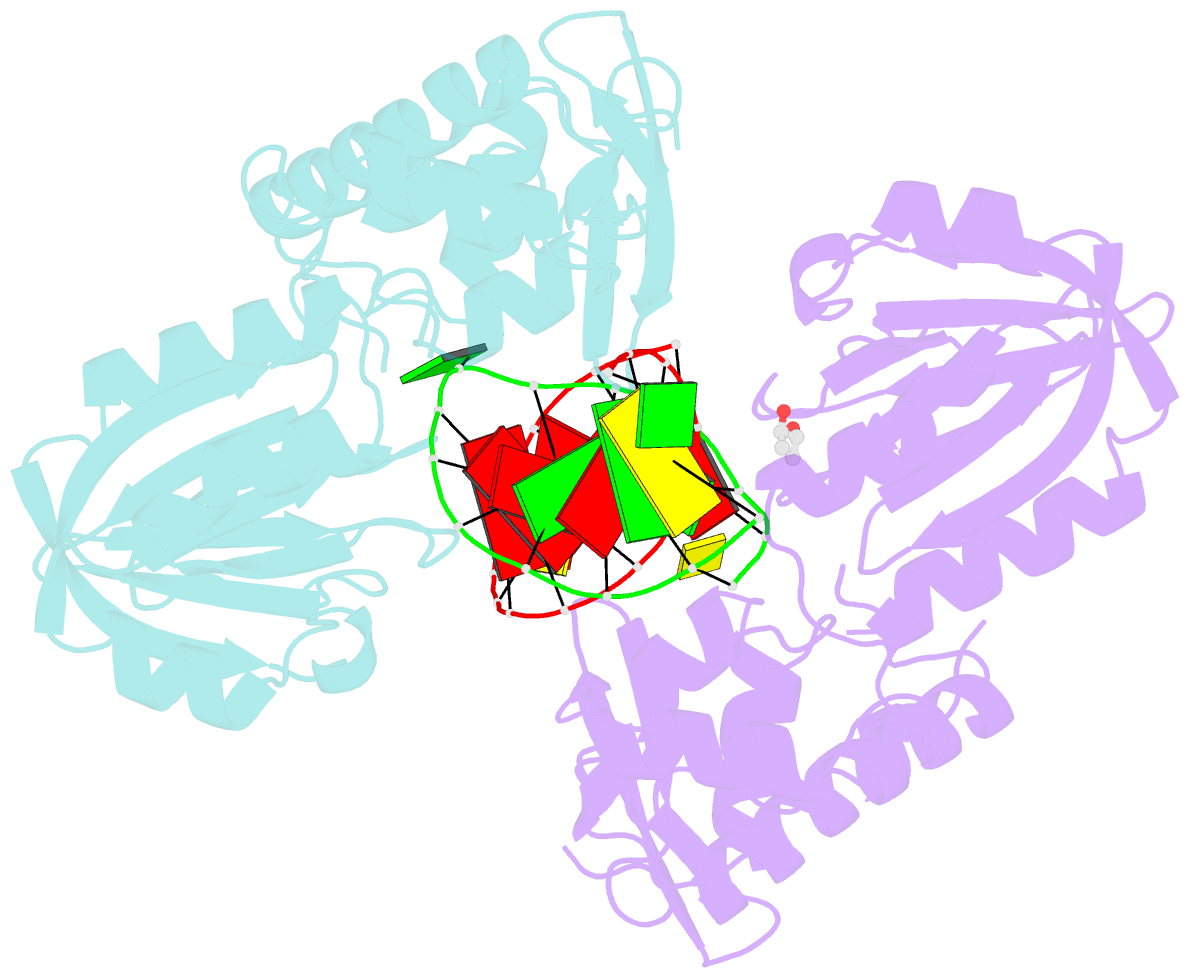
Summary information and primary citation
- PDB-id
- 4cis; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.05 Å)
- Summary
- Structure of mutm in complex with carbocyclic 8-oxo-g containing DNA
- Reference
- Sadeghian K, Flaig D, Blank ID, Schneider S, Strasser R, Stathis D, Winnacker M, Carell T, Ochsenfeld C (2014): "Ribose-protonated DNA base excision repair: a combined theoretical and experimental study." Angew. Chem. Int. Ed. Engl., 53, 10044-10048. doi: 10.1002/anie.201403334.
- Abstract
- Living organisms protect the genome against external influences by recognizing and repairing damaged DNA. A common source of gene mutation is the oxidized guanine, which undergoes base excision repair through cleavage of the glycosidic bond between the ribose and the nucleobase of the lesion. We unravel the repair mechanism utilized by bacterial glycosylase, MutM, using quantum-chemical calculations involving more than 1000 atoms of the catalytic site. In contrast to the base-protonated pathway currently favored in the literature, we show that the initial protonation of the lesion's ribose paves the way for an almost barrier-free glycosidic cleavage. The combination of theoretical and experimental data provides further insight into the selectivity and discrimination of MutM's binding site toward various substrates.