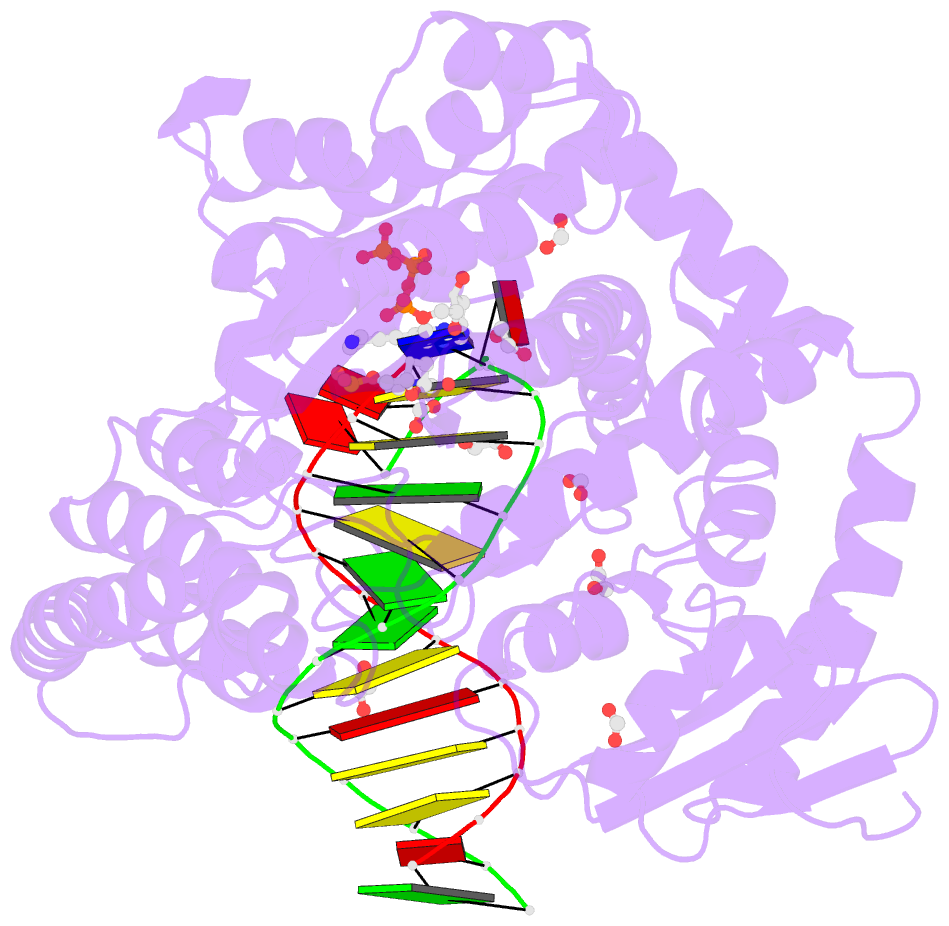
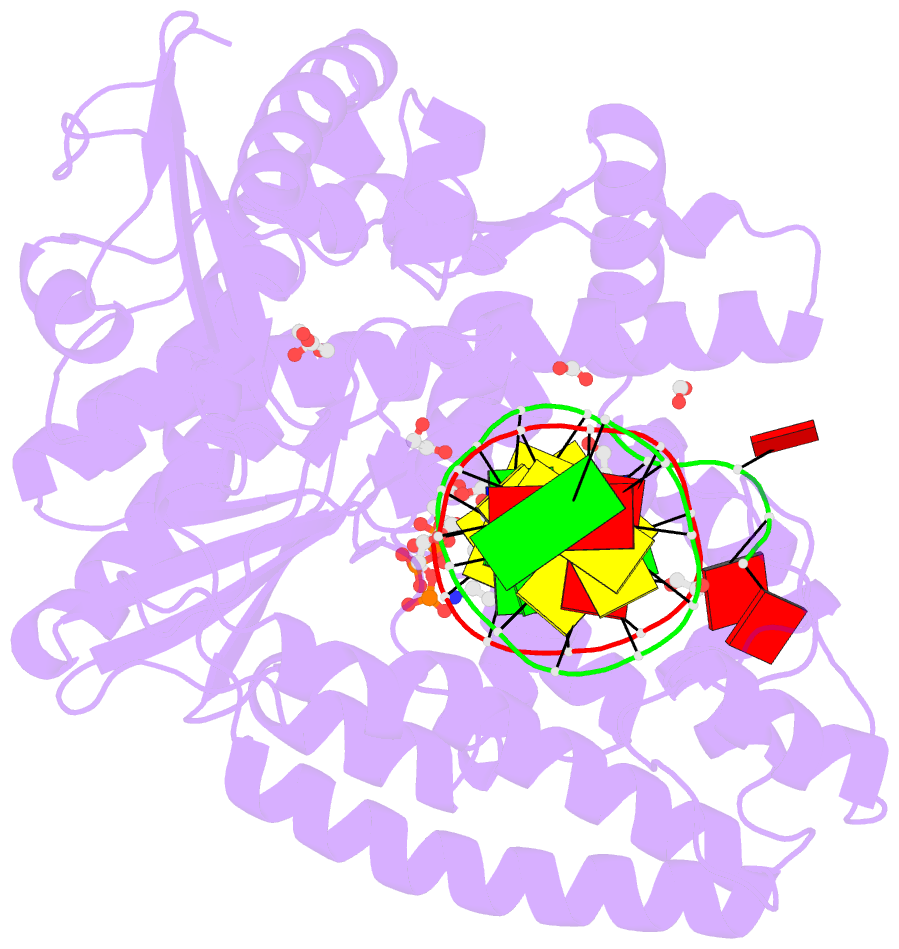
Summary information and primary citation
- PDB-id
- 4df8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.0 Å)
- Summary
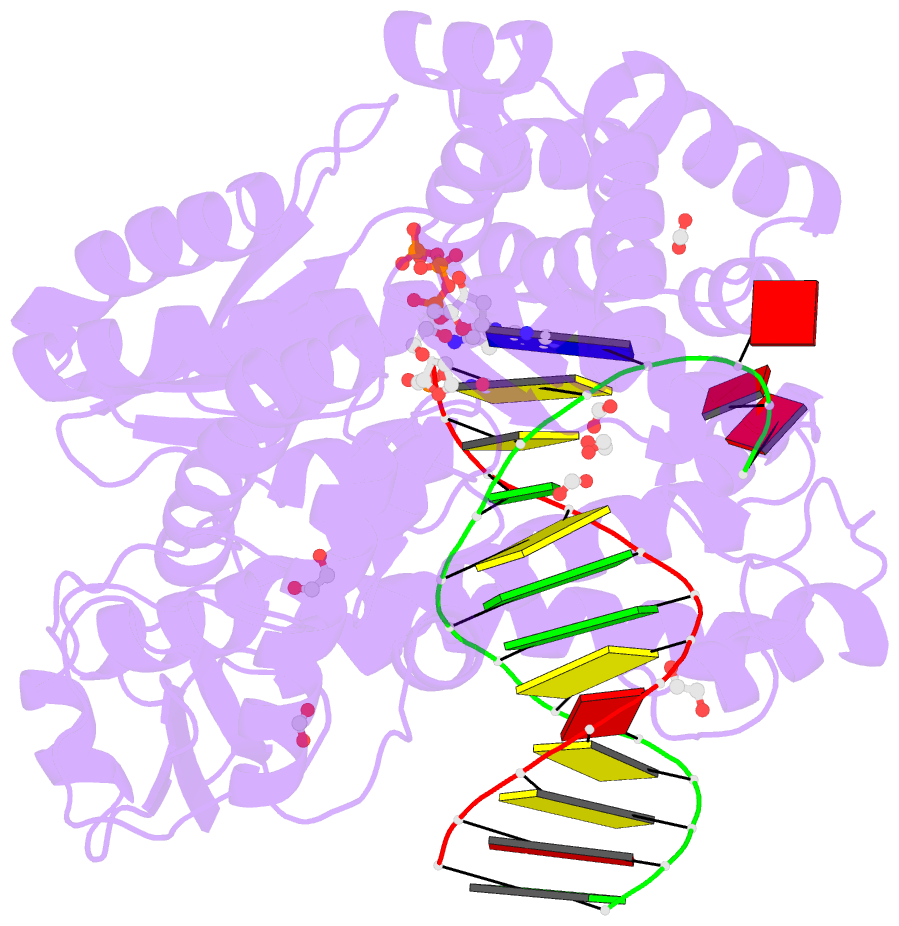
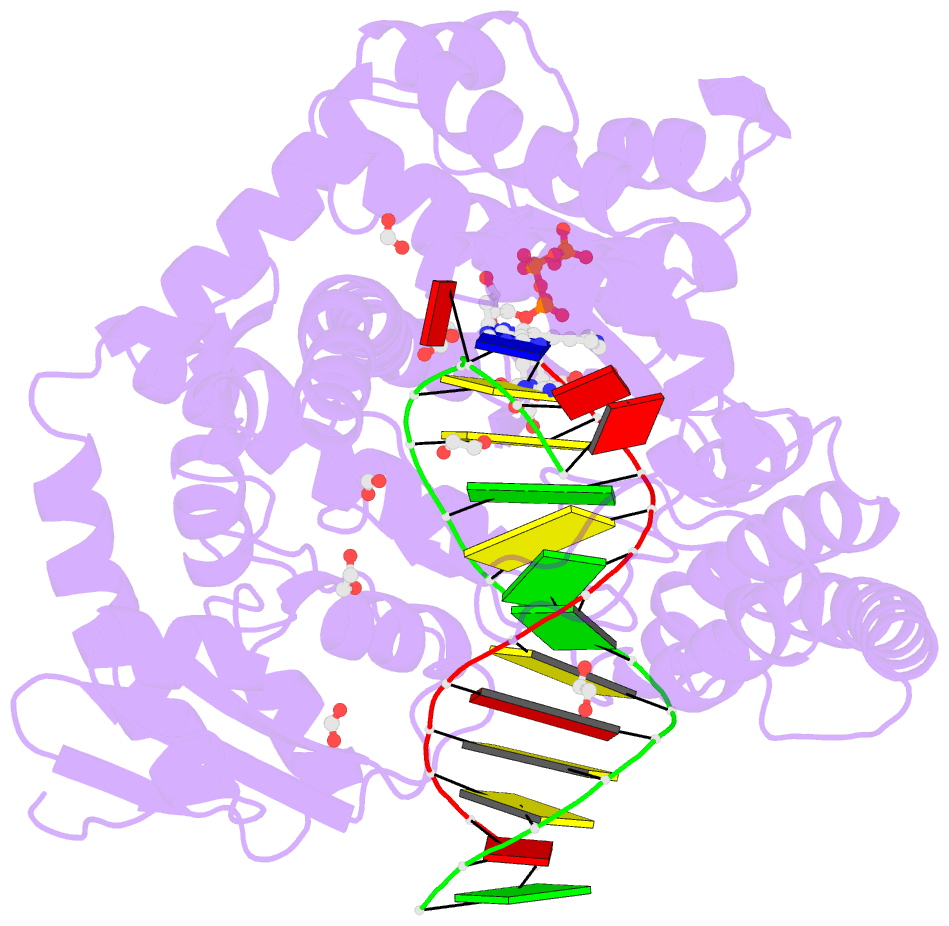
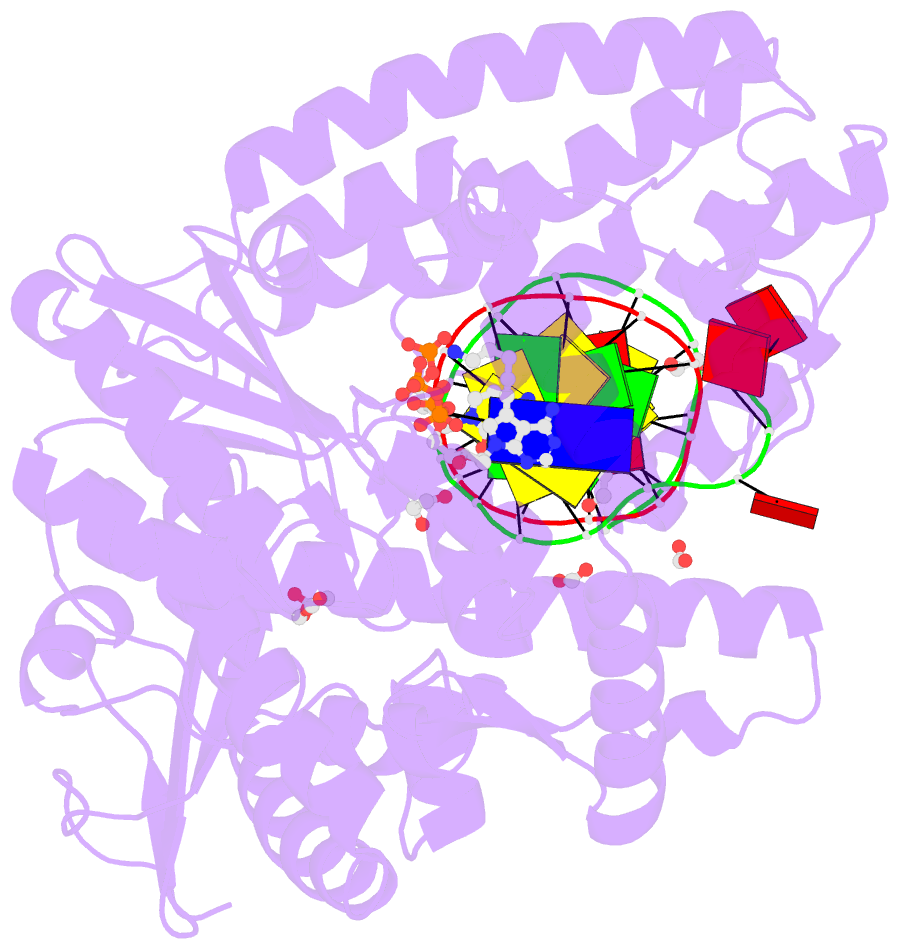
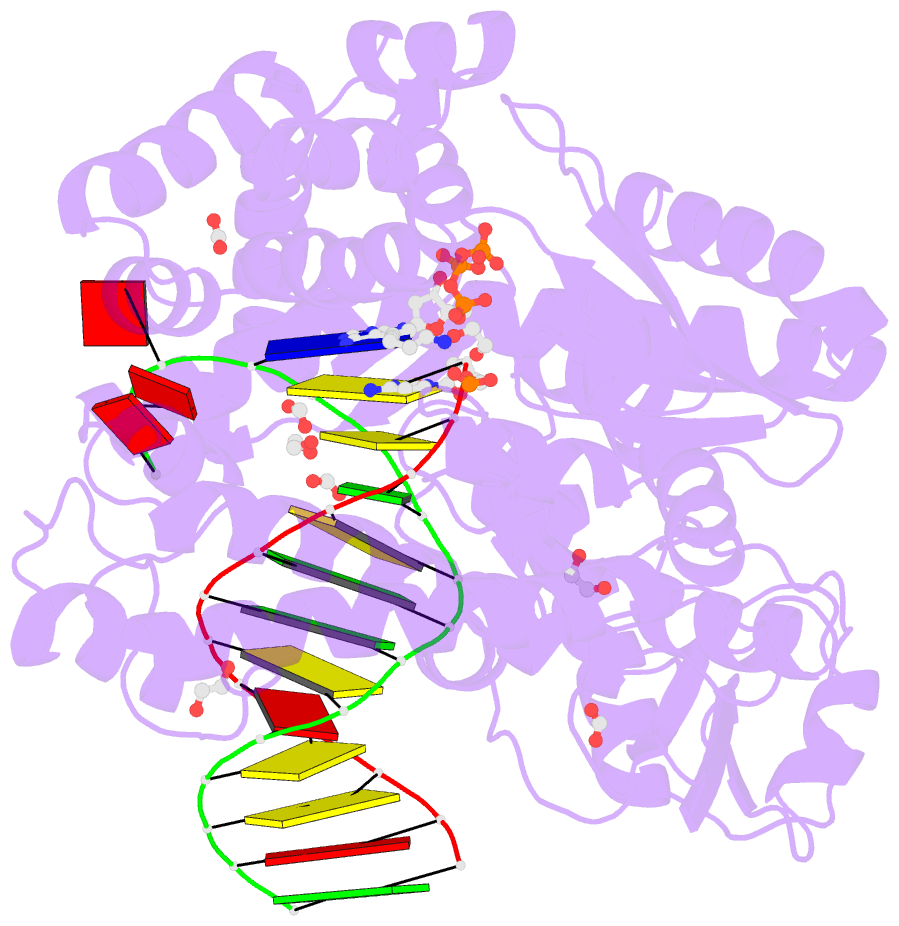
- Crystal structure of the large fragment of DNA polymerase i from thermus aquaticus in a closed ternary complex with aminopentinyl-7-deaza-2-datp
- Reference
- Bergen K, Steck AL, Strutt S, Baccaro A, Welte W, Diederichs K, Marx A (2012): "Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates." J.Am.Chem.Soc., 134, 11840-11843. doi: 10.1021/ja3017889.
- Abstract
- The capability of DNA polymerases to accept chemically modified nucleotides is of paramount importance for many biotechnological applications. Although these analogues are widely used, the structural basis for the acceptance of the unnatural nucleotide surrogates has been only sparsely explored. Here we present in total six crystal structures of modified 2'-deoxynucleoside-5'-O-triphosphates (dNTPs) carrying modifications at the C5 positions of pyrimidines or C7 positions of 7-deazapurines in complex with a DNA polymerase and a primer/template complex. The modified dNTPs are in positions poised for catalysis leading to incorporation. These structural data provide insight into the mechanism of incorporation and acceptance of modified dNTPs. Our results open the door for rational design of modified nucleotides, which should offer great opportunities for future applications.