Summary information and primary citation
- PDB-id
- 4dl7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-inhibitor
- Method
- X-ray (1.97 Å)
- Summary
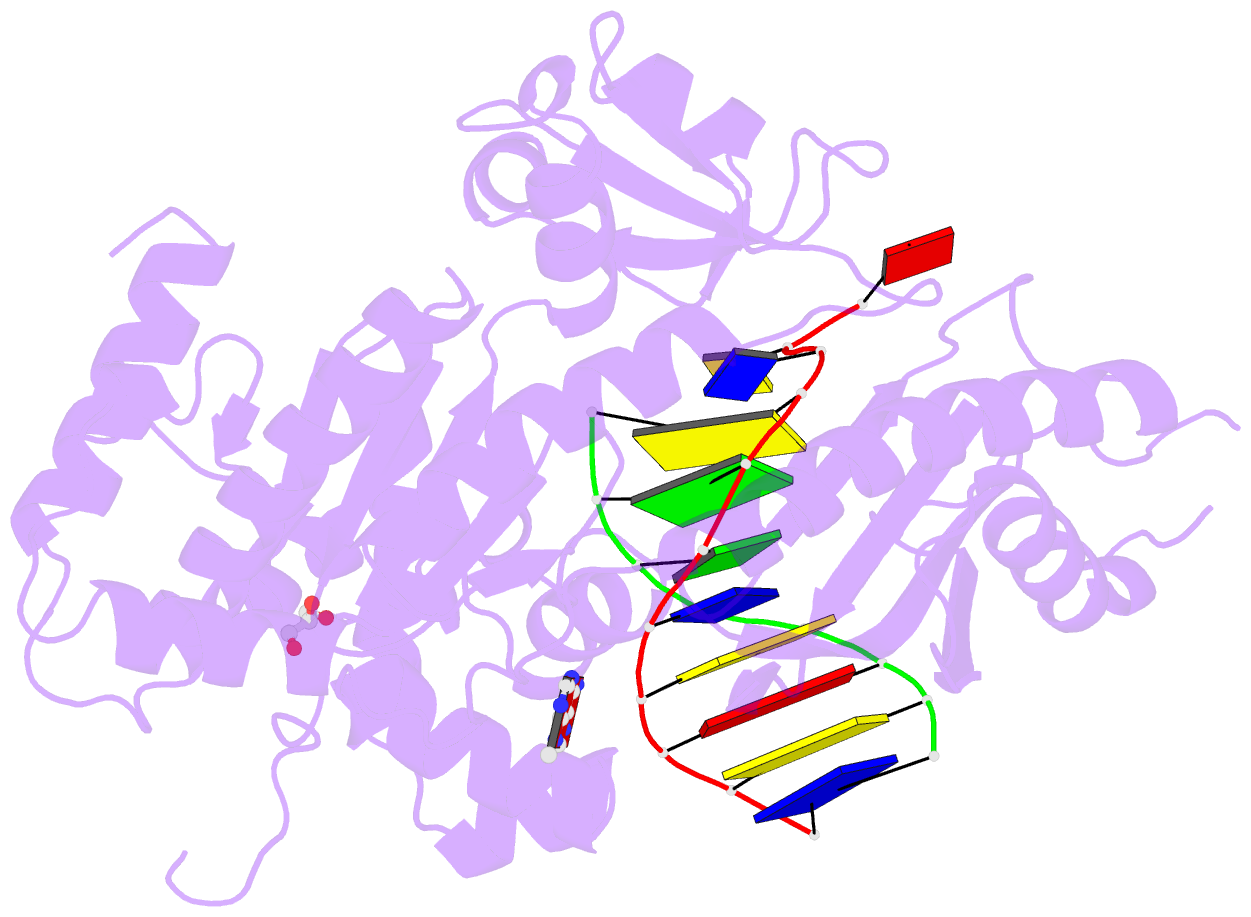
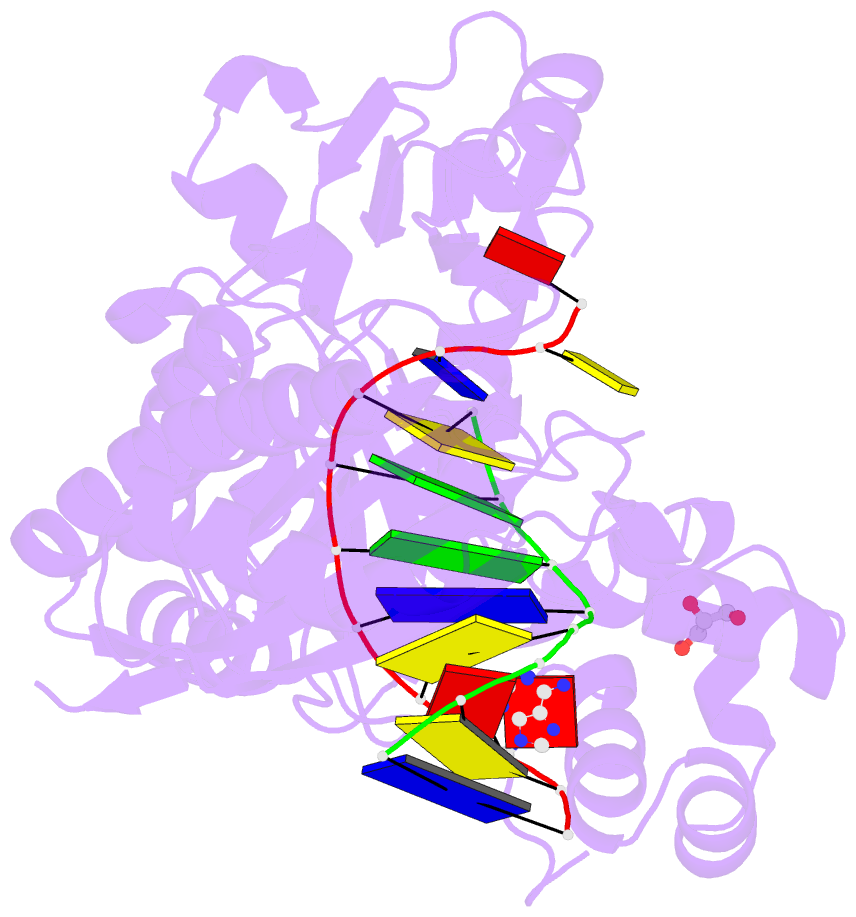
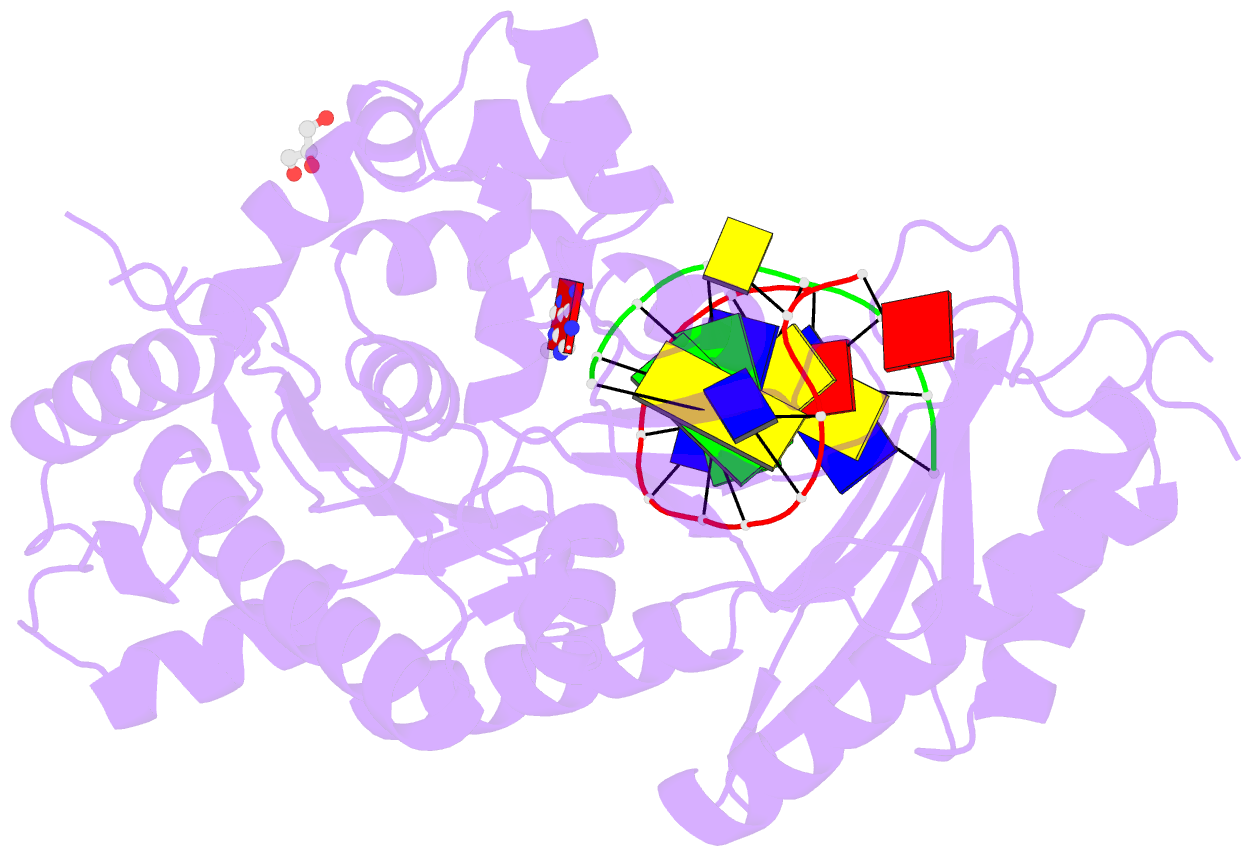
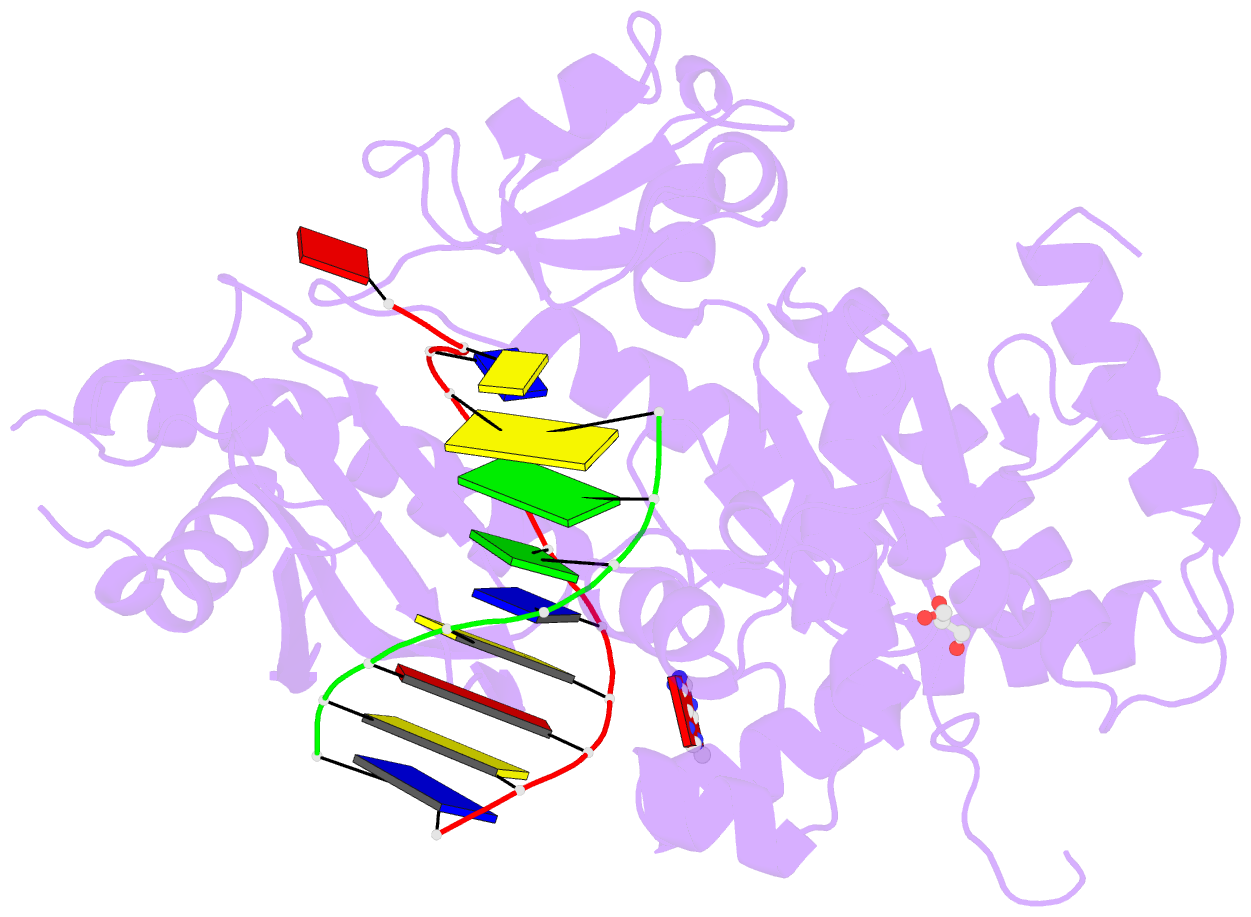
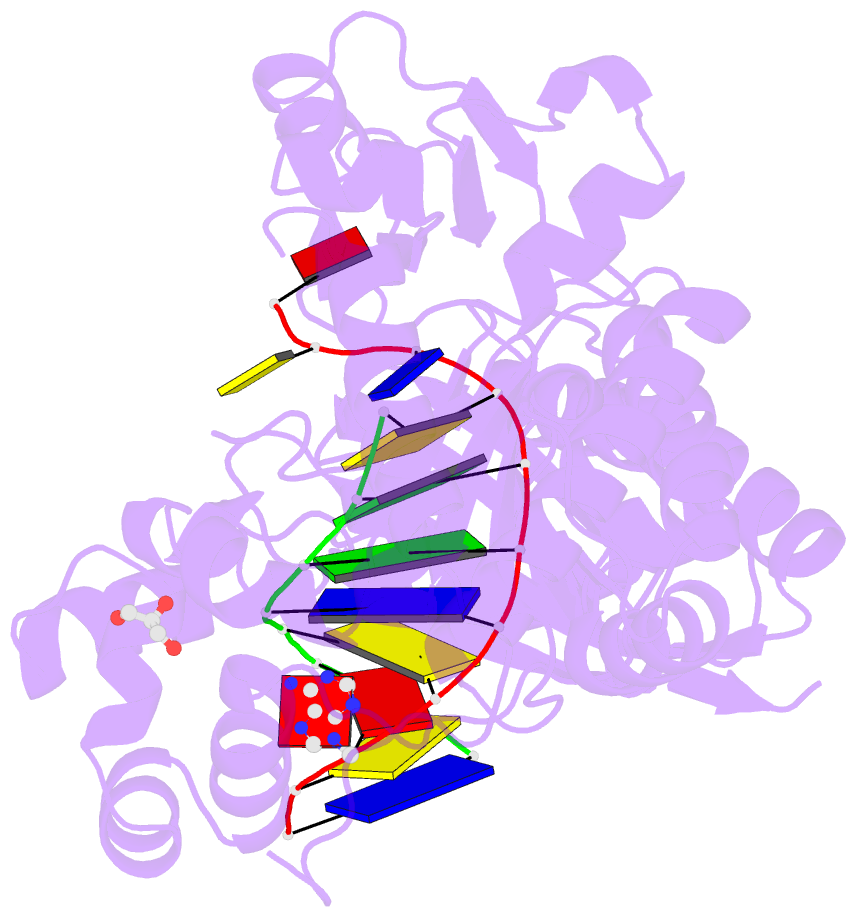
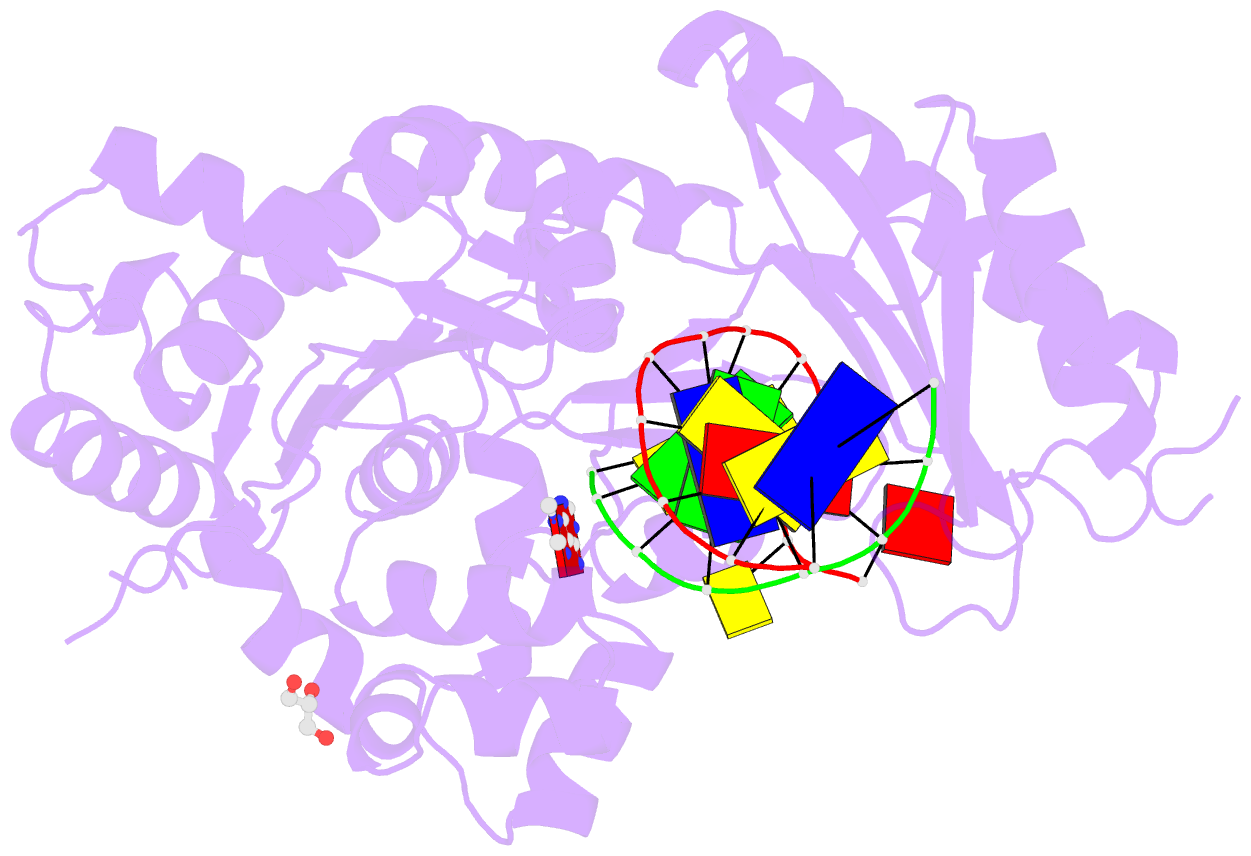
- Human DNA polymerase eta fails to extend primer 2 nucleotide after cisplatin crosslink (pt-gg4).
- Reference
- Zhao Y, Biertumpfel C, Gregory MT, Hua YJ, Hanaoka F, Yang W (2012): "Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta." Proc.Natl.Acad.Sci.USA, 109, 7269-7274. doi: 10.1073/pnas.1202681109.
- Abstract
- Cisplatin (cis-diamminedichloroplatinum) and related compounds cause DNA damage and are widely used as anticancer agents. Chemoresistance to cisplatin treatment is due in part to translesion synthesis by human DNA polymerase η (hPol η). Here, we report crystal structures of hPol η complexed with intrastrand cisplatin-1,2-cross-linked DNA, representing four consecutive steps in translesion synthesis. In contrast to the generally enlarged and nondiscriminating active site of Y-family polymerases like Dpo4, Pol η is specialized for efficient bypass of UV-cross-linked pyrimidine dimers. Human Pol η differs from the yeast homolog in its binding of DNA template. To incorporate deoxycytidine opposite cisplatin-cross-linked guanines, hPol η undergoes a specific backbone rearrangement to accommodate the larger base dimer and minimizes the DNA distortion around the lesion. Our structural analyses show why Pol η is inefficient at extending primers after cisplatin lesions, which necessitates a second translesion DNA polymerase to complete bypass in vivo. A hydrophobic pocket near the primer-binding site in human Pol η is identified as a potential drug target for inhibiting translesion synthesis and, thereby, reducing chemoresistance.