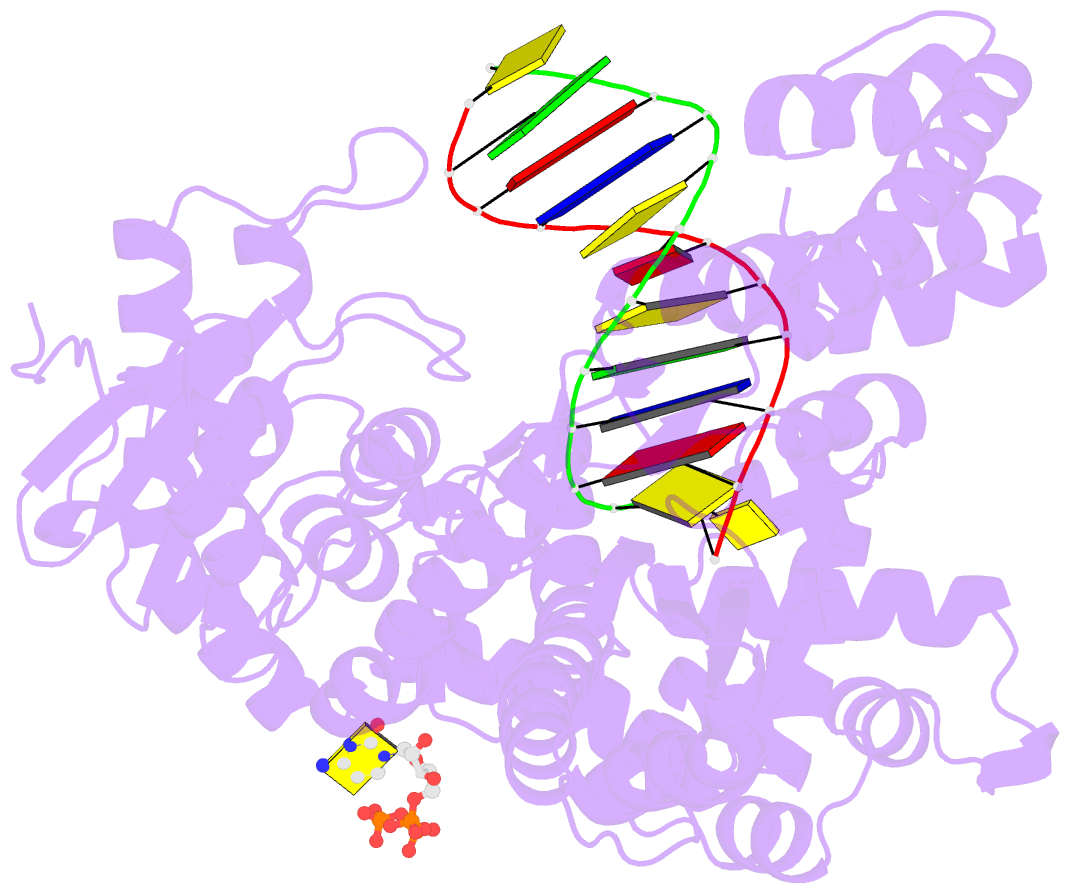
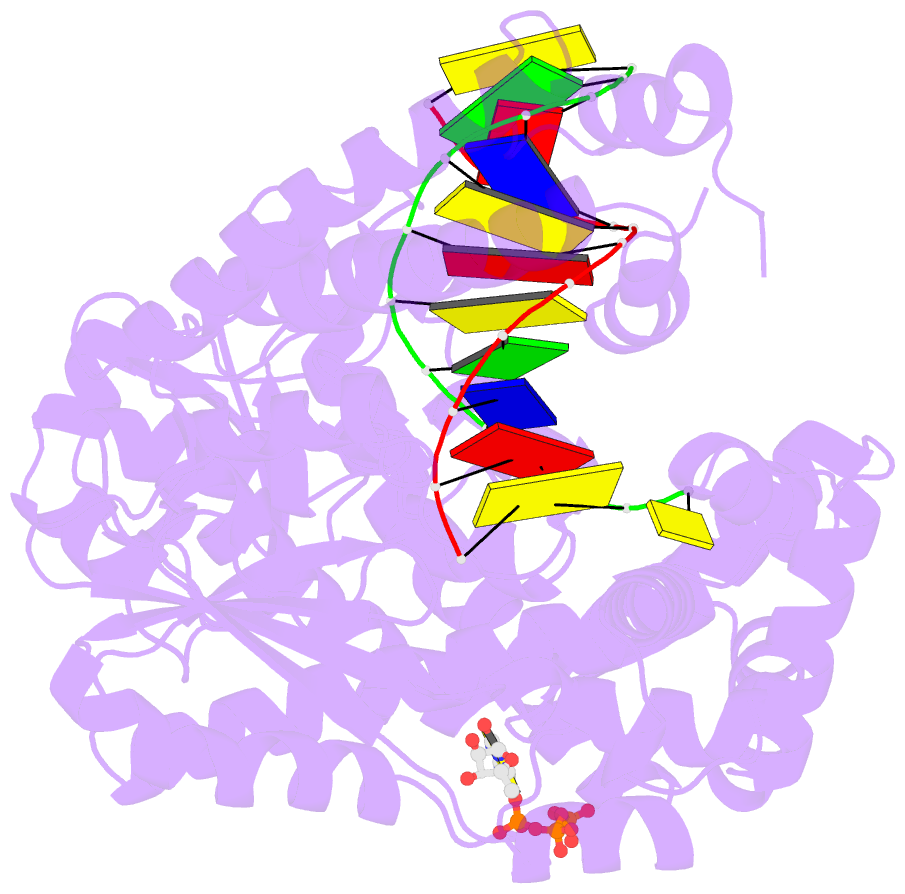
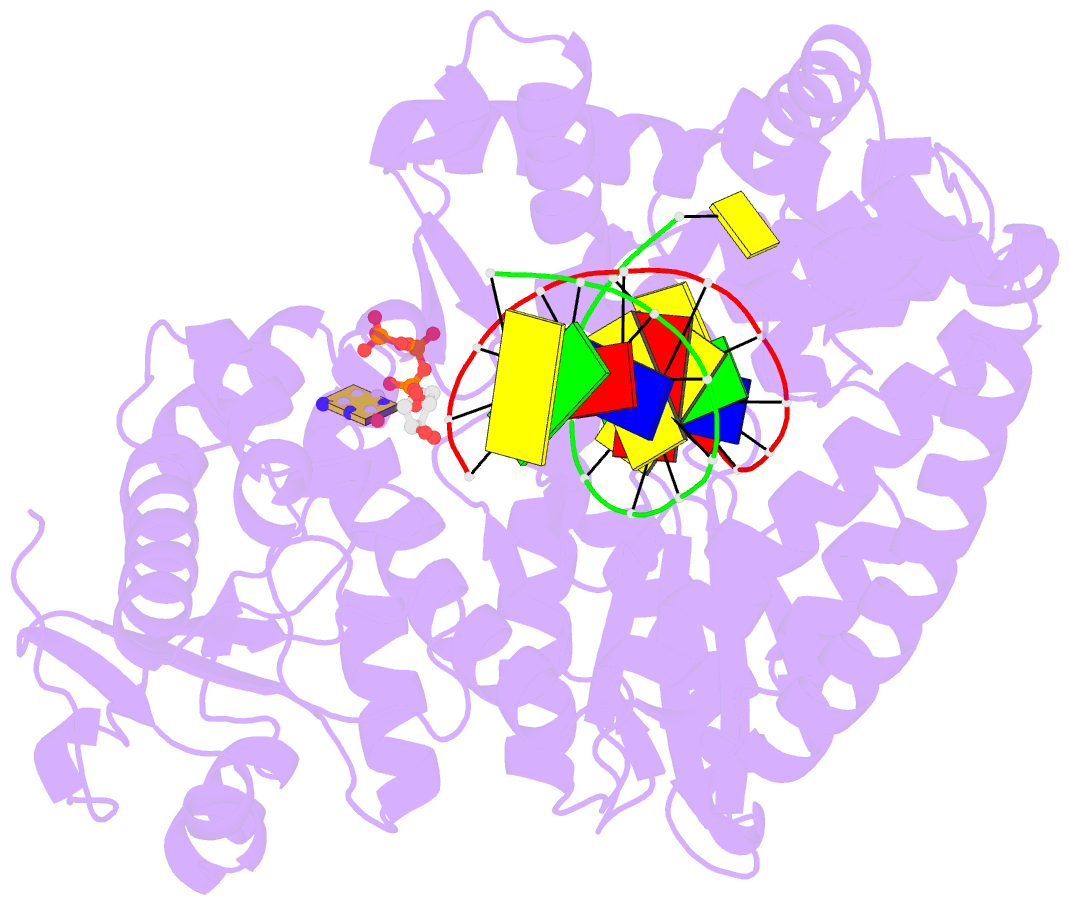
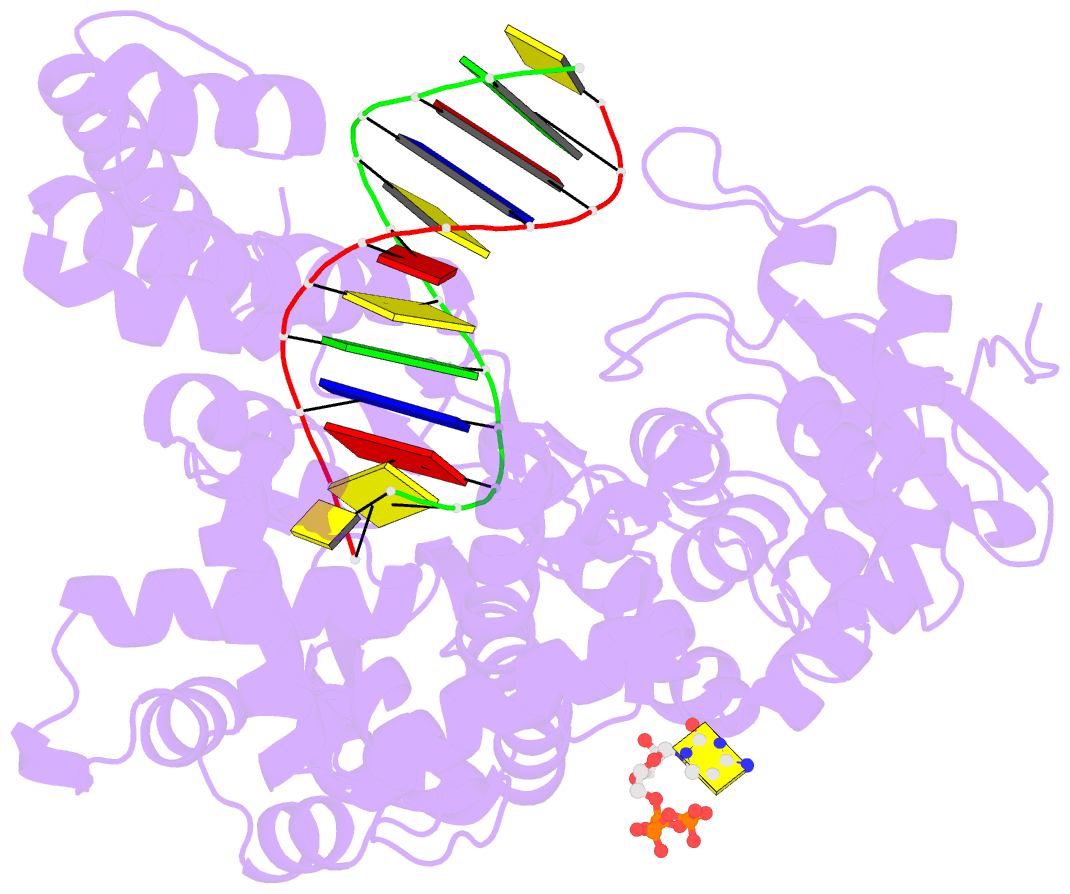
Summary information and primary citation
- PDB-id
- 4dqs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.66 Å)
- Summary
- Binary complex of bacillus DNA polymerase i large fragment and duplex DNA with rc in primer terminus paired with dg of template
- Reference
- Wang W, Wu EY, Hellinga HW, Beese LS (2012): "Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides." J.Biol.Chem., 287, 28215-28226. doi: 10.1074/jbc.M112.366609.
- Abstract
- In addition to discriminating against base pair mismatches, DNA polymerases exhibit a high degree of selectivity for deoxyribonucleotides over ribo- or dideoxynucleotides. It has been proposed that a single active site residue (steric gate) blocks productive binding of nucleotides containing 2'-hydroxyls. Although this steric gate plays a role in sugar moiety discrimination, its interactions do not account fully for the observed behavior of mutants. Here we present 10 high resolution crystal structures and enzyme kinetic analyses of Bacillus DNA polymerase I large fragment variants complexed with deoxy-, ribo-, and dideoxynucleotides and a DNA substrate. Taken together, these data present a more nuanced and general mechanism for nucleotide discrimination in which ensembles of intermediate conformations in the active site trap non-cognate substrates. It is known that the active site O-helix transitions from an open state in the absence of nucleotide substrates to a ternary complex closed state in which the reactive groups are aligned for catalysis. Substrate misalignment in the closed state plays a fundamental part in preventing non-cognate nucleotide misincorpation. The structures presented here show that additional O-helix conformations intermediate between the open and closed state extremes create an ensemble of binding sites that trap and misalign non-cognate nucleotides. Water-mediated interactions, absent in the fully closed state, play an important role in formation of these binding sites and can be remodeled to accommodate different non-cognate substrates. This mechanism may extend also to base pair discrimination.