Summary information and primary citation
- PDB-id
- 4dw7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (3.08 Å)
- Summary
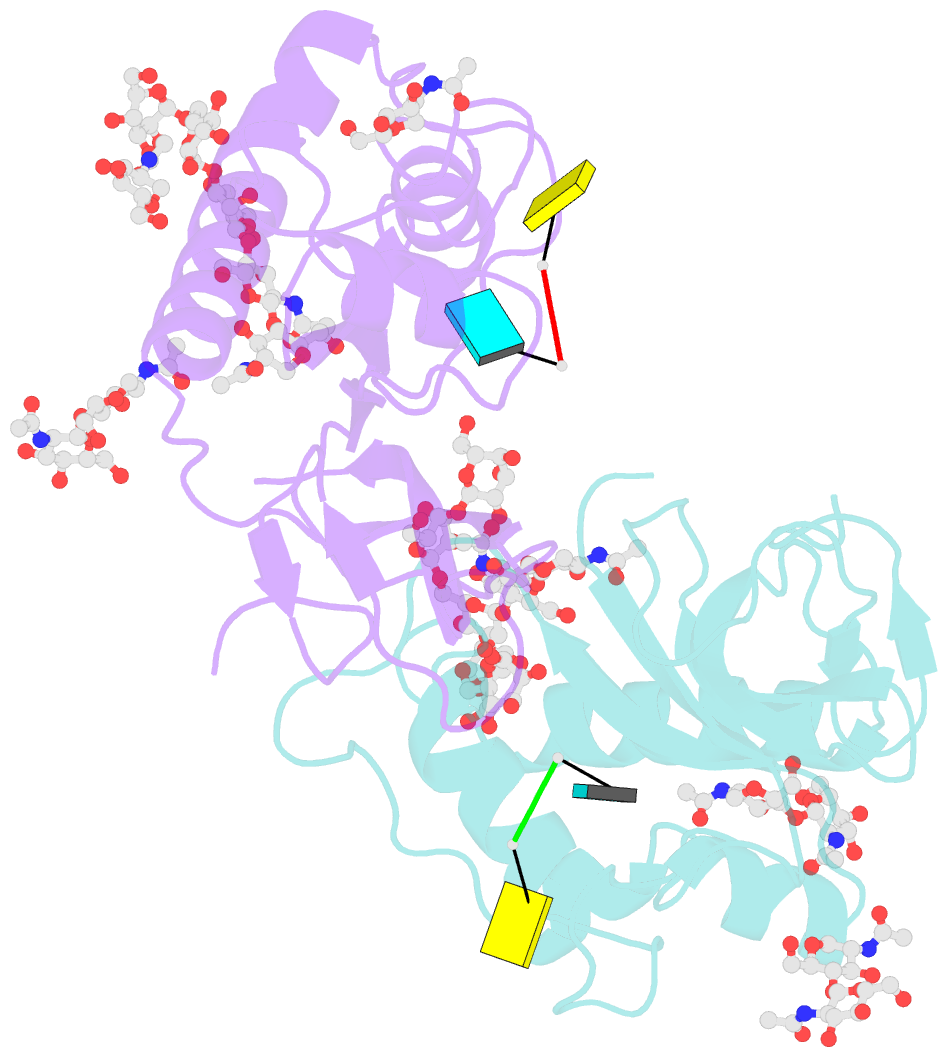
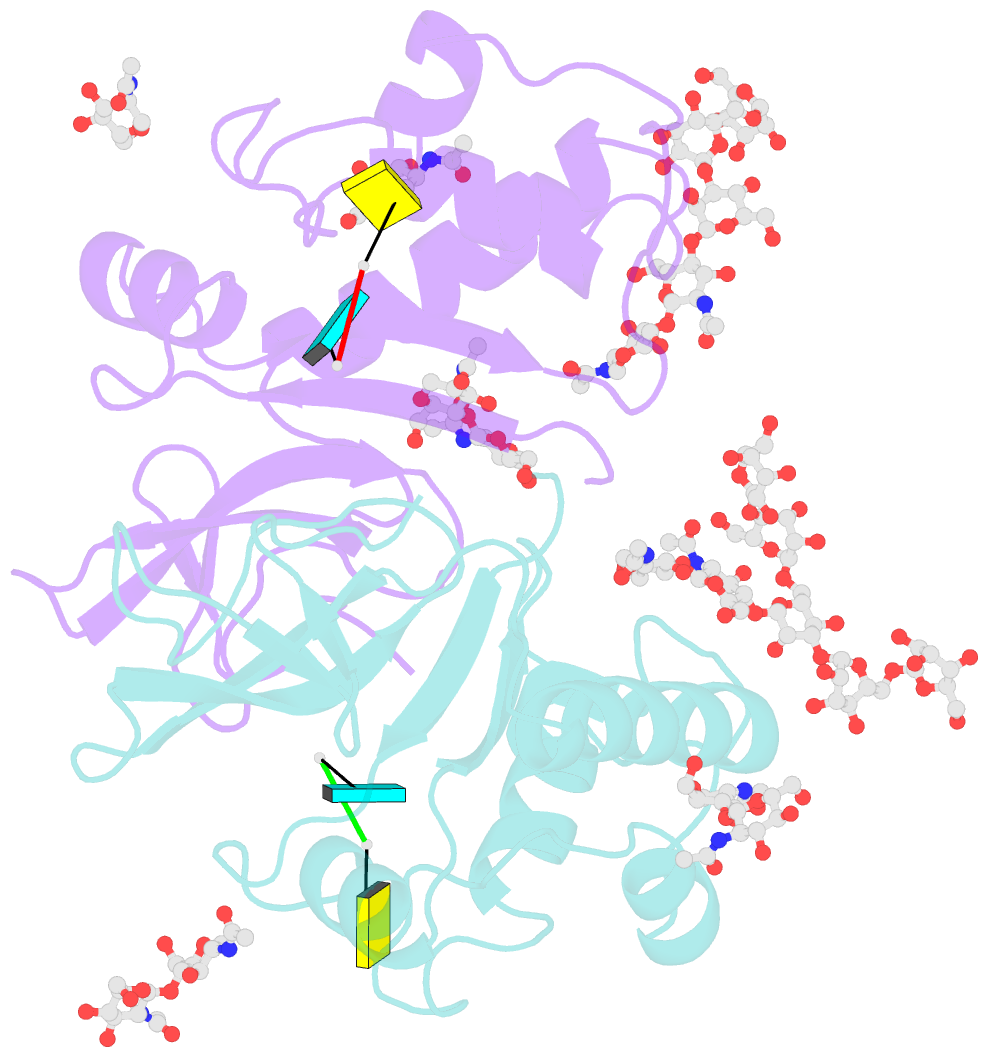
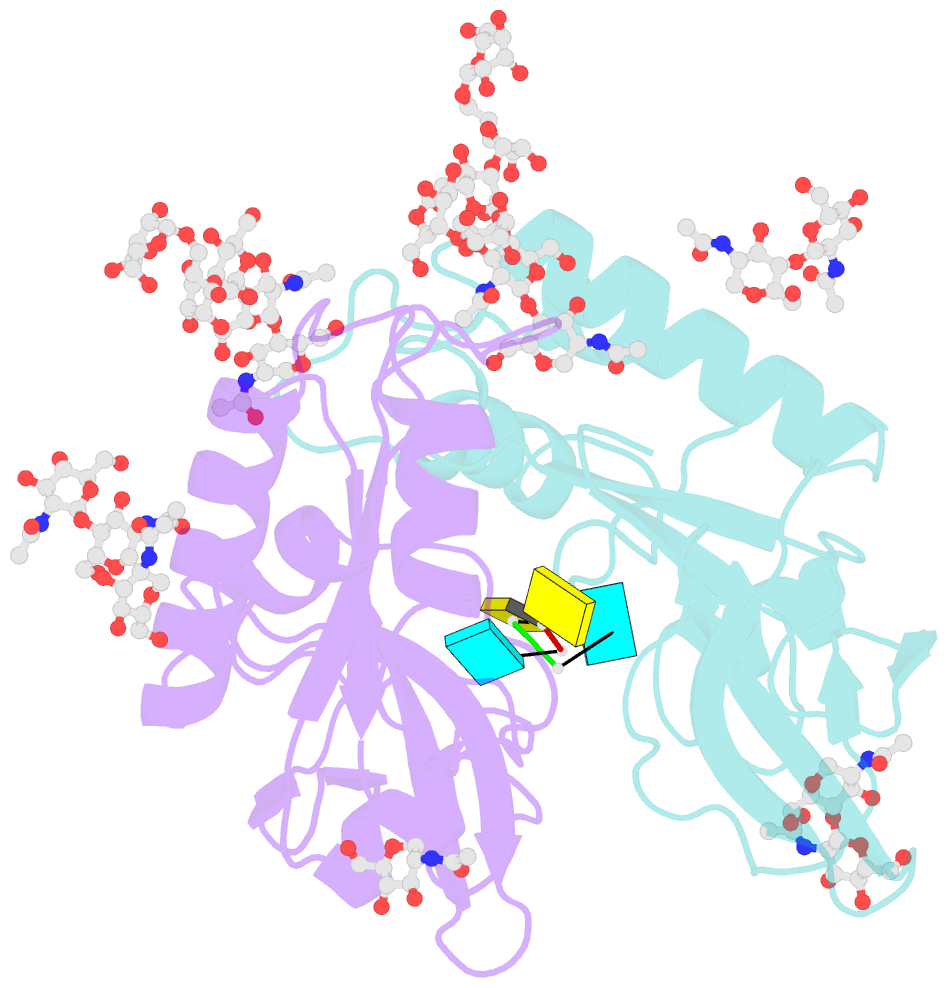
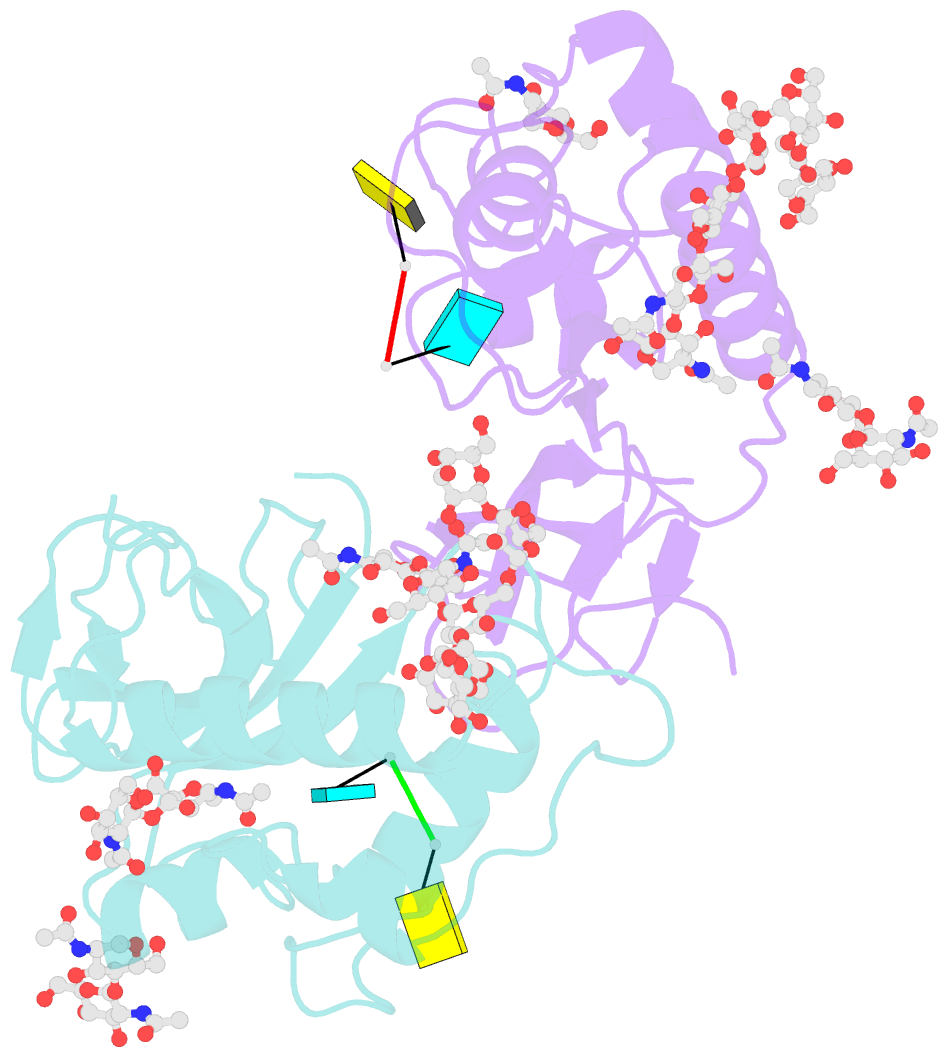
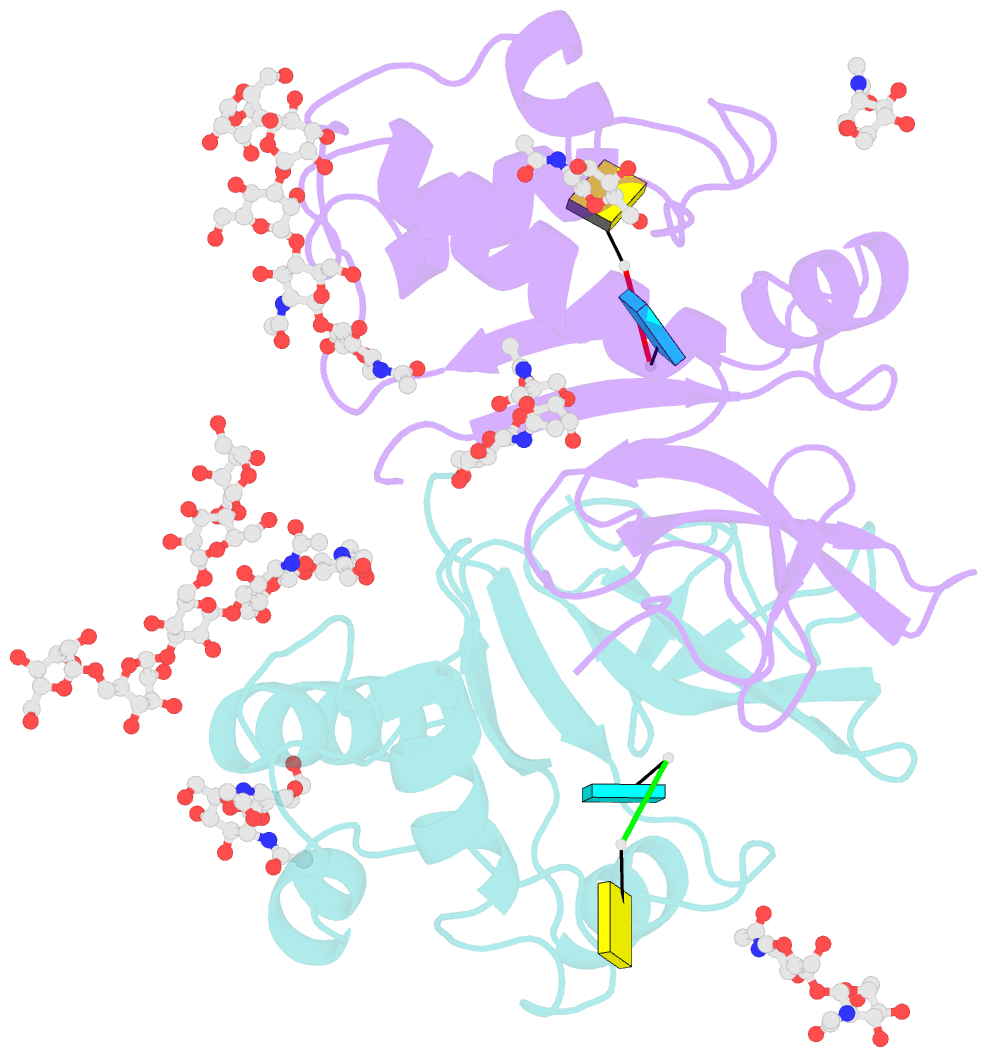
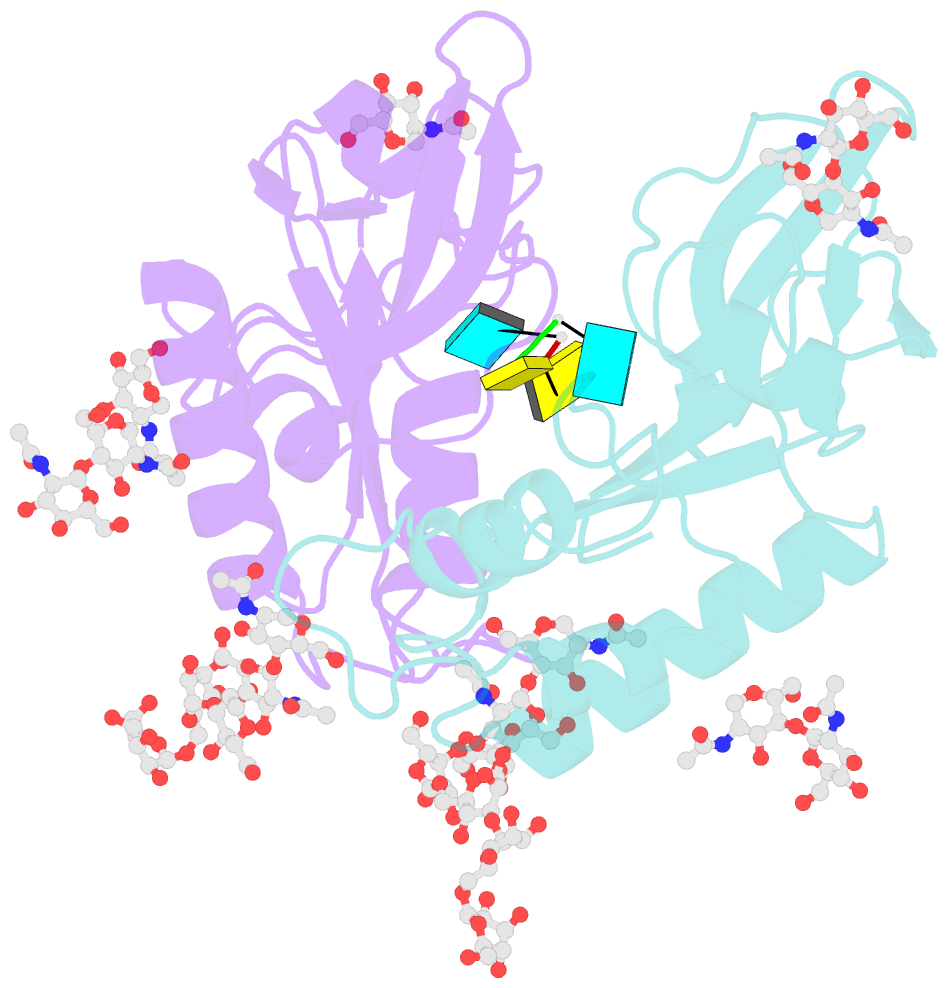
- Crystal structure of an active-site mutant of the glycoprotein erns from the pestivirus bvdv-1 in complex with a cpu dinucleotide
- Reference
- Krey T, Bontems F, Vonrhein C, Vaney MC, Bricogne G, Rumenapf T, Rey FA (2012): "Crystal Structure of the Pestivirus Envelope Glycoprotein E(rns) and Mechanistic Analysis of Its Ribonuclease Activity." Structure, 20, 862-873. doi: 10.1016/j.str.2012.03.018.
- Abstract
- Pestiviruses, which belong to the Flaviviridae family of RNA viruses, are important agents of veterinary diseases causing substantial economical losses in animal farming worldwide. Pestivirus particles display three envelope glycoproteins at their surface: E(rns), E1, and E2. We report here the crystal structure of the catalytic domain of E(rns), the ribonucleolytic activity of which is believed to counteract the innate immunity of the host. The structure reveals a three-dimensional fold corresponding to T2 ribonucleases from plants and fungi. Cocrystallization experiments with mono- and oligonucleotides revealed the structural basis for substrate recognition at two binding sites previously identified for T2 RNases. A detailed analysis of poly-U cleavage products using (31)P-NMR and size exclusion chromatography, together with molecular docking studies, provides a comprehensive mechanistic picture of E(rns) activity on its substrates and reveals the presence of at least one additional nucleotide binding site.