Summary information and primary citation
- PDB-id
- 4egz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.3 Å)
- Summary
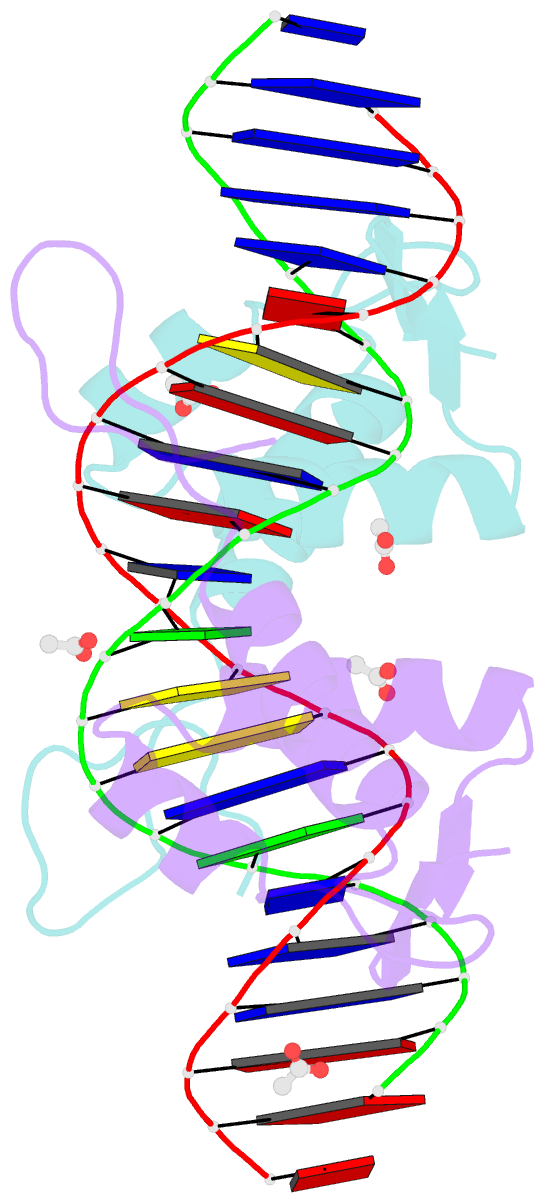
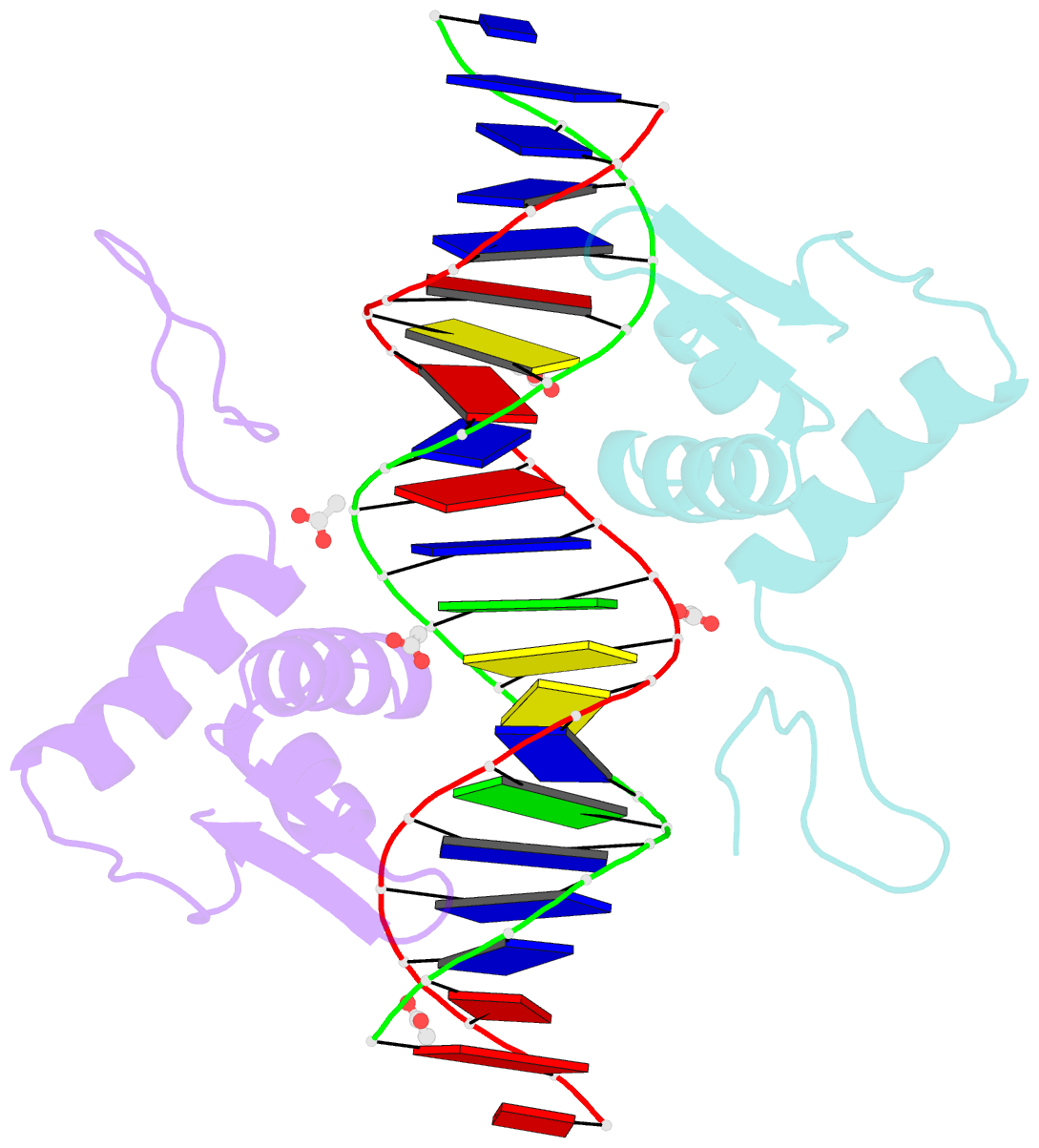
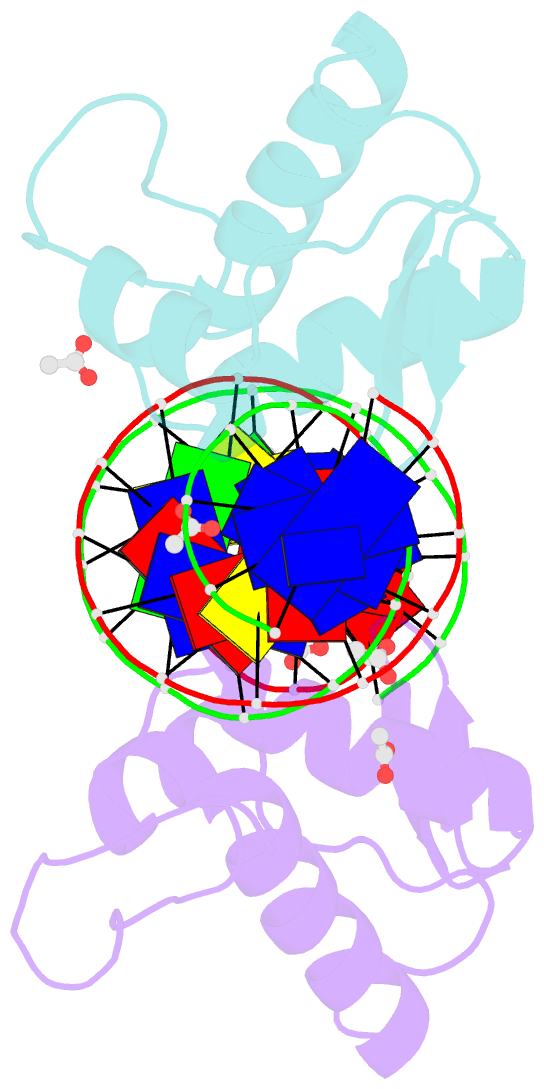
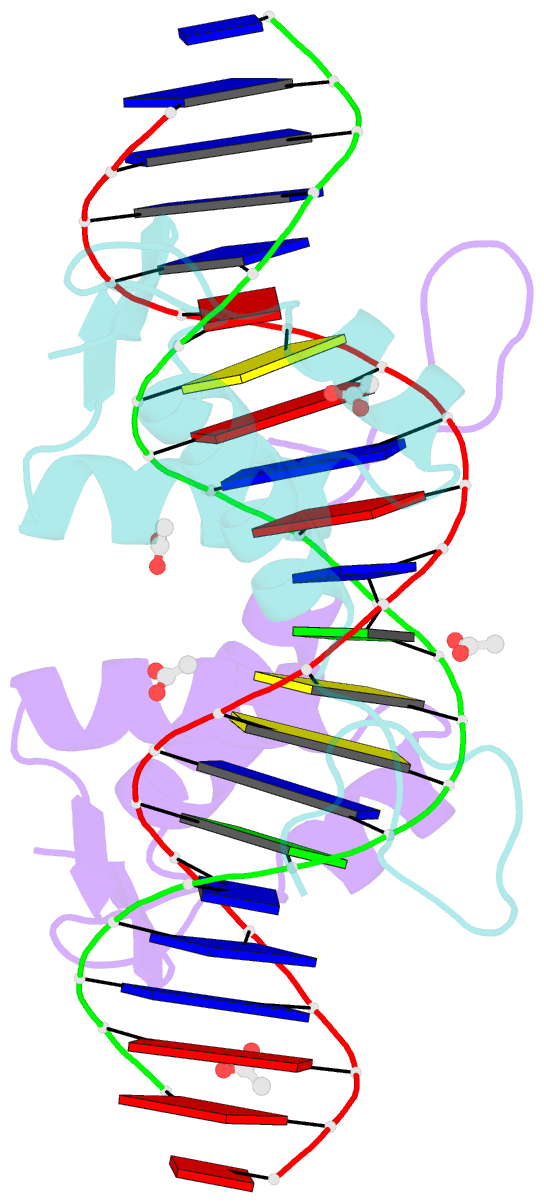
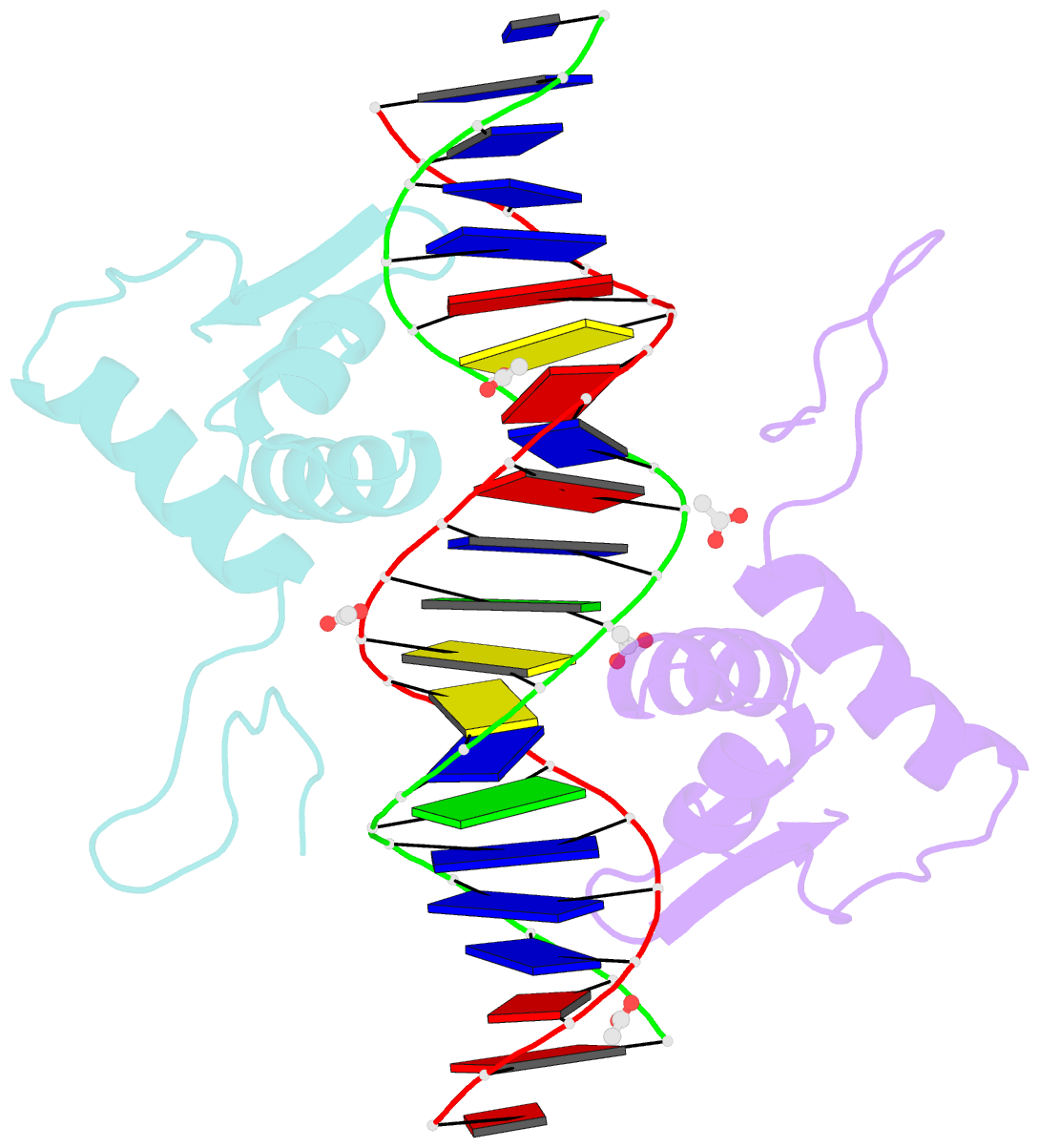
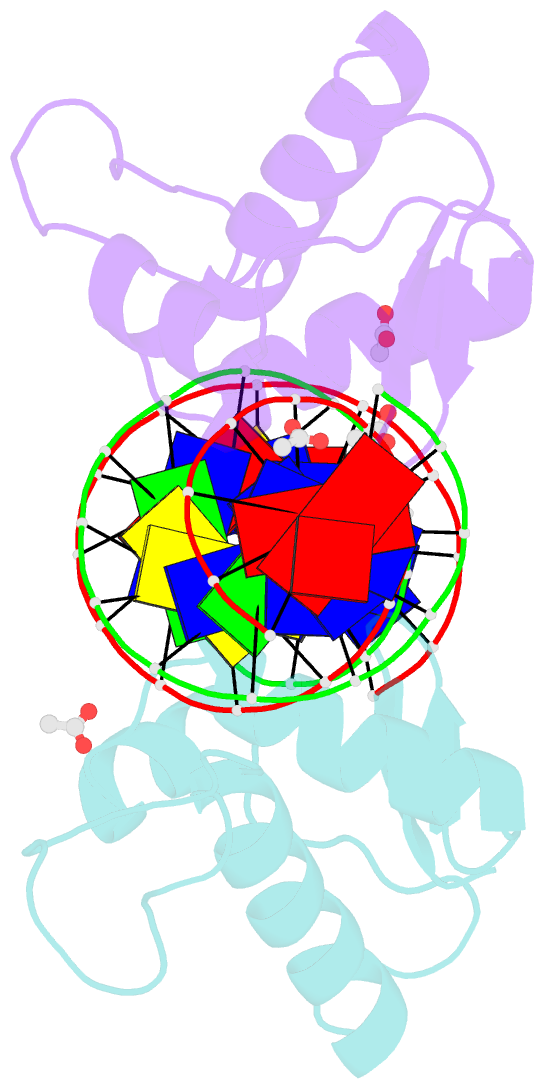
- Crystal structure of arar(dbd) in complex with operator orr3
- Reference
- Jain D, Nair DT (2013): "Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators." Nucleic Acids Res., 41, 639-647. doi: 10.1093/nar/gks962.
- Abstract
- Transcription factors modulate expression primarily through specific recognition of cognate sequences resident in the promoter region of target genes. AraR (Bacillus subtilis) is a repressor of genes involved in L-arabinose metabolism. It binds to eight different operators present in five different promoters with distinct affinities through a DNA binding domain at the N-terminus. The structures of AraR-NTD in complex with two distinct operators (ORA1 and ORR3) reveal that two monomers bind to one recognition motif (T/ANG) each in the bipartite operators. The structures show that the two recognition motifs are spaced apart by six bases in cases of ORA1 and eight bases in case of ORR3. This increase in the spacing in the operators by two base pairs results in a drastic change in the position and orientation of the second monomer on DNA in the case of ORR3 when compared with ORA1. Because AraR binds to the two operators with distinct affinities to achieve different levels of repression, this observation suggests that the variation in the spacing between core recognition motifs could be a strategy used by this transcription modulator to differentially influence gene expression.