Summary information and primary citation
- PDB-id
- 4f3t; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.25 Å)
- Summary
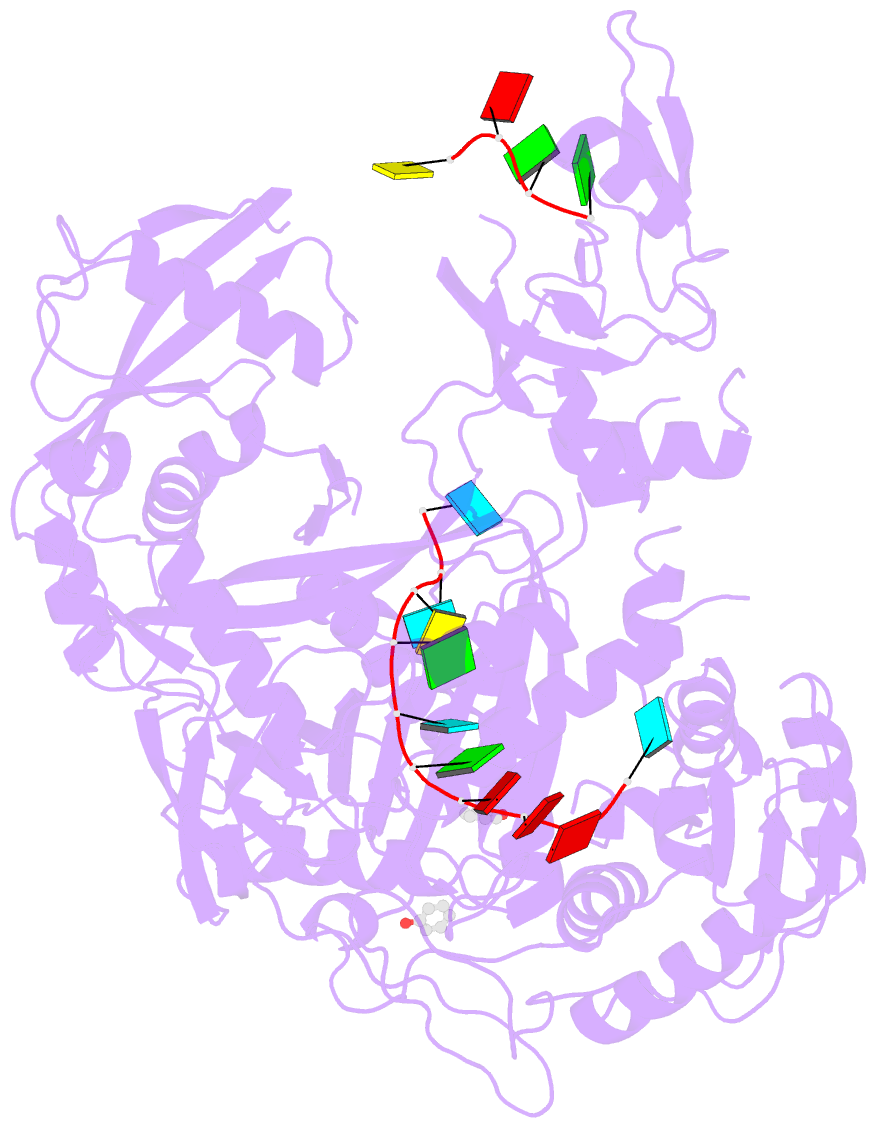
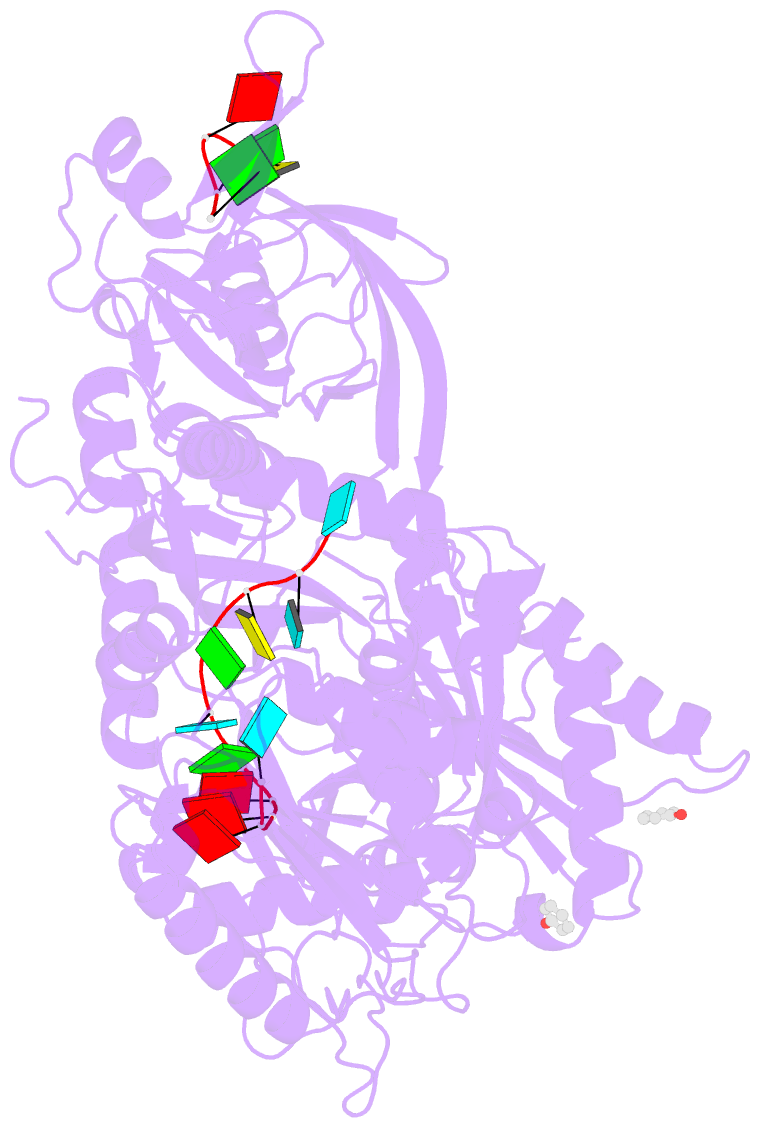
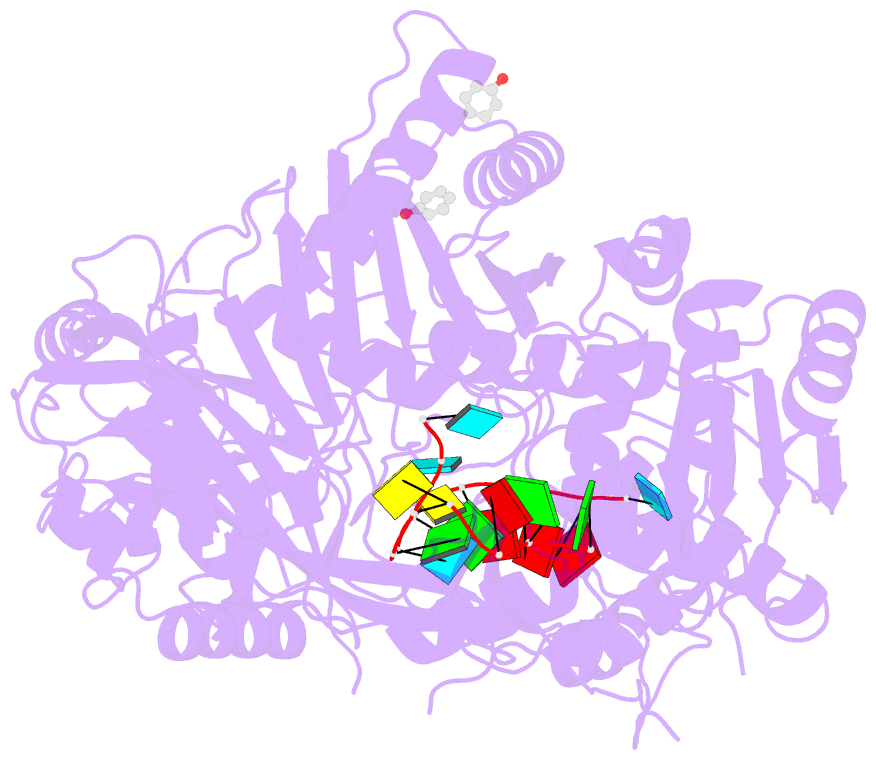
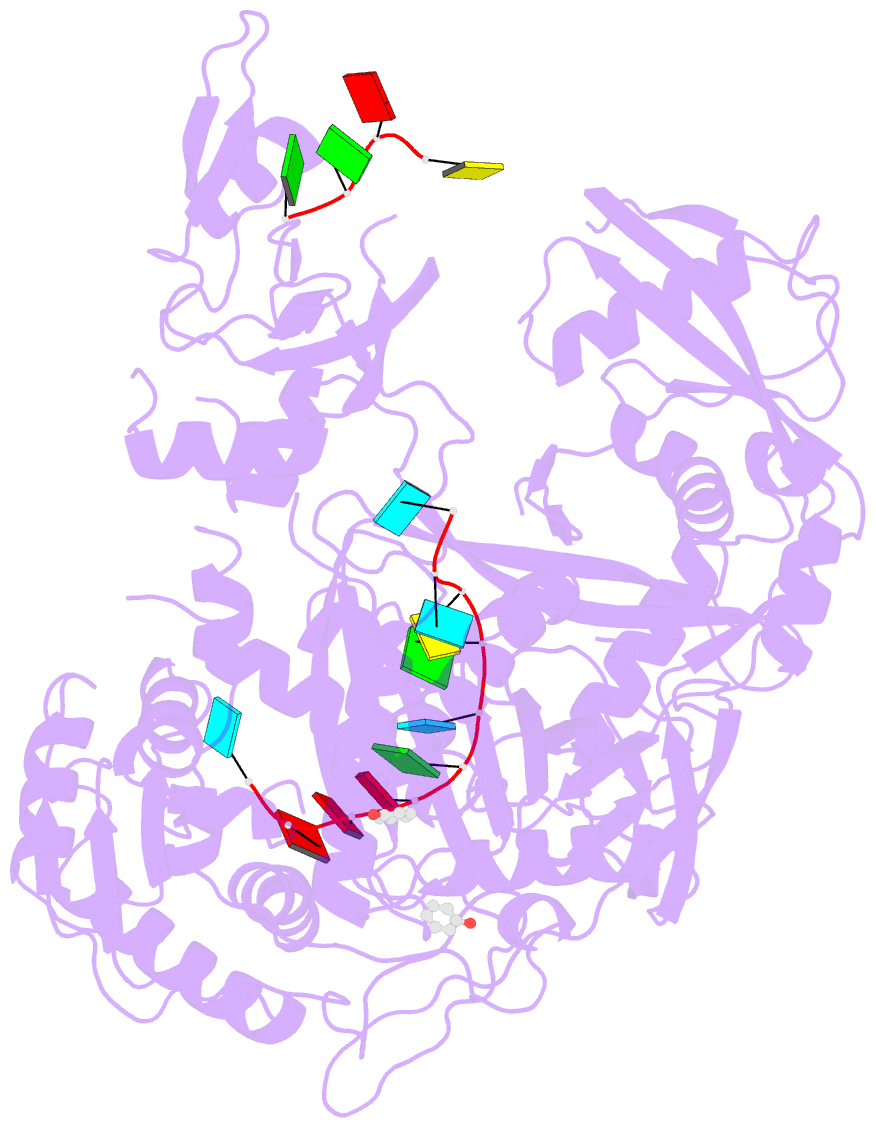
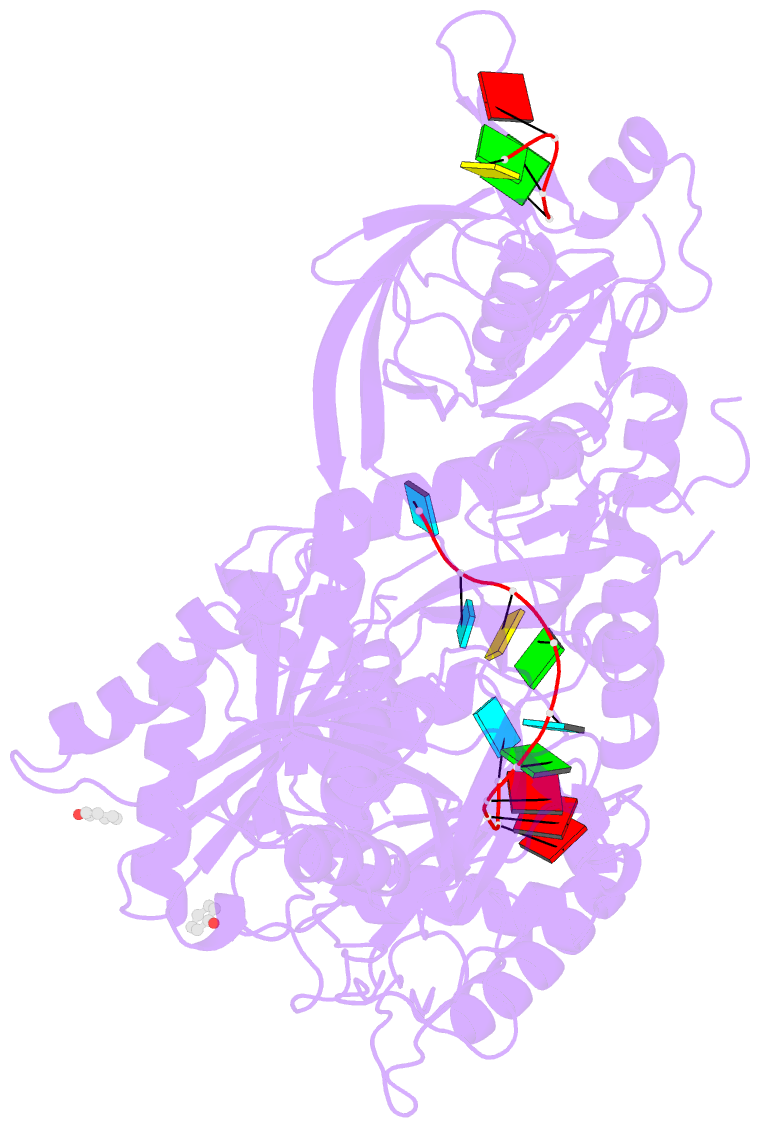
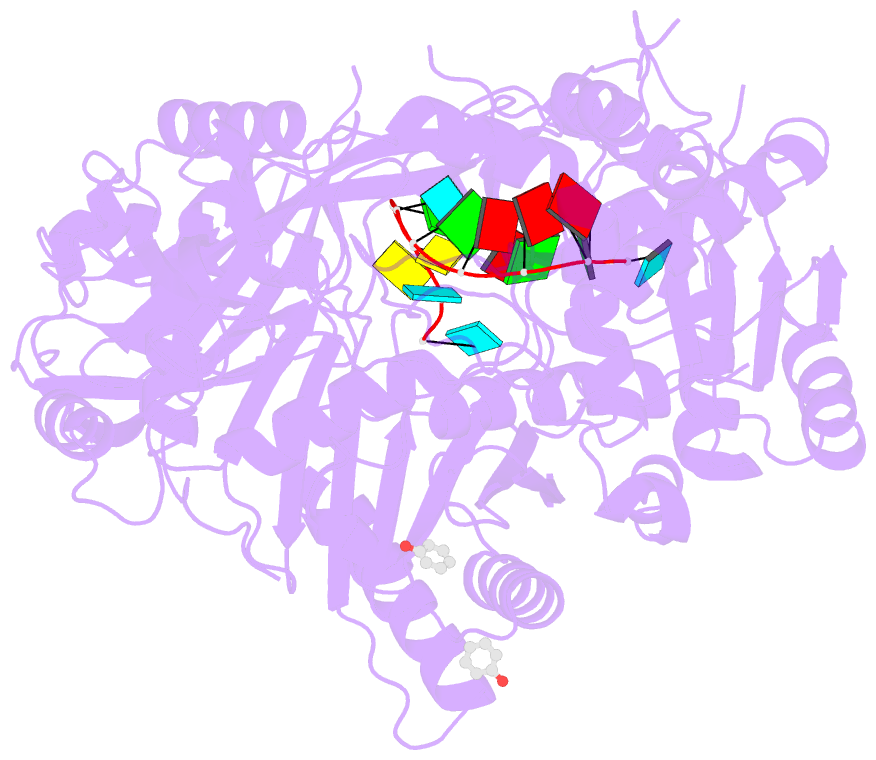
- Human argonaute-2 - mir-20a complex
- Reference
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L (2012): "The Structure of Human Argonaute-2 in Complex with miR-20a." Cell(Cambridge,Mass.), 150, 100-110. doi: 10.1016/j.cell.2012.05.017.
- Abstract
- Argonaute proteins lie at the heart of the RNA-induced silencing complex (RISC), wherein they use small RNA guides to recognize targets. Initial insight into the architecture of Argonautes came from studies of prokaryotic proteins, revealing a crescent-shaped base made up of the amino-terminal, PAZ, middle, and PIWI domains. The recently reported crystal structure of human Argonaute-2 (hAgo2), the "slicer" in RNA interference, in complex with a mixed population of RNAs derived from insect cells provides insight into the architecture of a eukaryotic Argonaute protein with defined biochemical and biological functions. Here, we report the structure of human Ago2 bound to a physiologically relevant microRNA, microRNA-20a, at 2.2 Å resolution. The miRNA is anchored at both ends by the Mid and PAZ domains and makes several kinks and turns along the binding groove. Interestingly, miRNA binding confers remarkable stability on hAgo2, locking this otherwise flexible enzyme into a stable conformation.