Summary information and primary citation
- PDB-id
- 4fcy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.706 Å)
- Summary
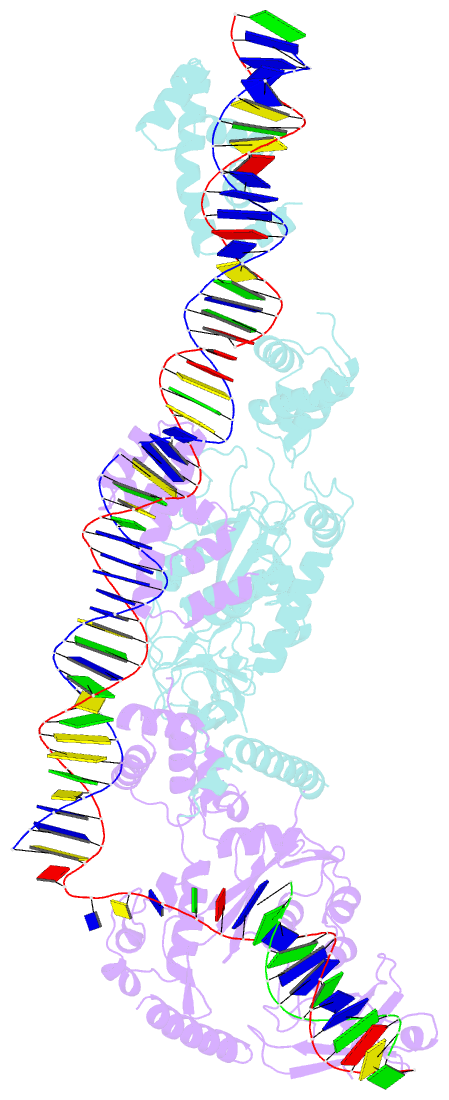
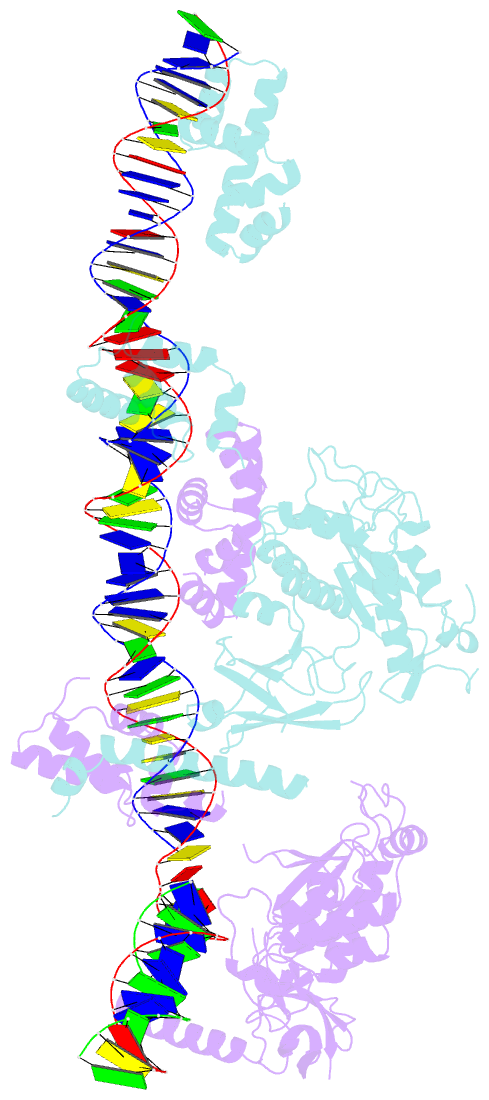
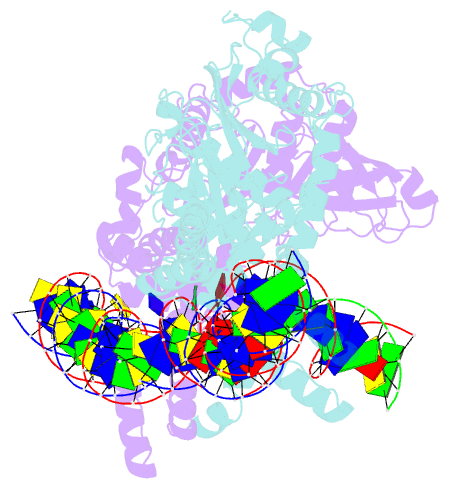
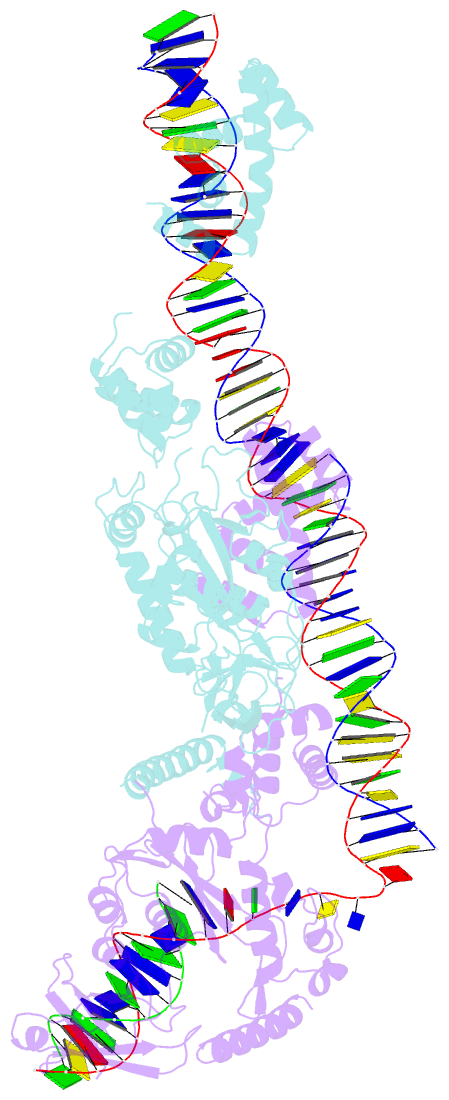
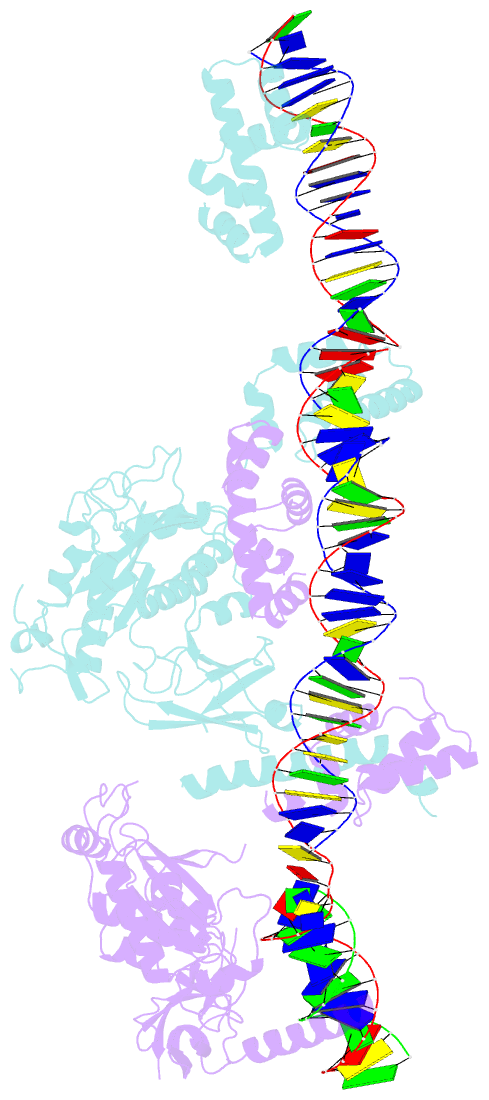
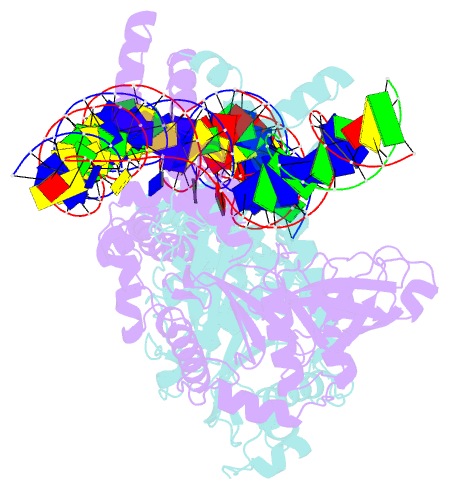
- Crystal structure of the bacteriophage mu transpososome
- Reference
- Montano SP, Pigli YZ, Rice PA (2012): "The Mu transpososome structure sheds light on DDE recombinase evolution." Nature, 491, 413-417. doi: 10.1038/nature11602.
- Abstract
- Studies of bacteriophage Mu transposition paved the way for understanding retroviral integration and V(D)J recombination as well as many other DNA transposition reactions. Here we report the structure of the Mu transpososome--Mu transposase (MuA) in complex with bacteriophage DNA ends and target DNA--determined from data that extend anisotropically to 5.2 Å, 5.2 Å and 3.7 Å resolution, in conjunction with previously determined structures of individual domains. The highly intertwined structure illustrates why chemical activity depends on formation of the synaptic complex, and reveals that individual domains have different roles when bound to different sites. The structure also provides explanations for the increased stability of the final product complex and for its preferential recognition by the ATP-dependent unfoldase ClpX. Although MuA and many other recombinases share a structurally conserved 'DDE' catalytic domain, comparisons among the limited set of available complex structures indicate that some conserved features, such as catalysis in trans and target DNA bending, arose through convergent evolution because they are important for function.