Summary information and primary citation
- PDB-id
- 4g0a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.1 Å)
- Summary
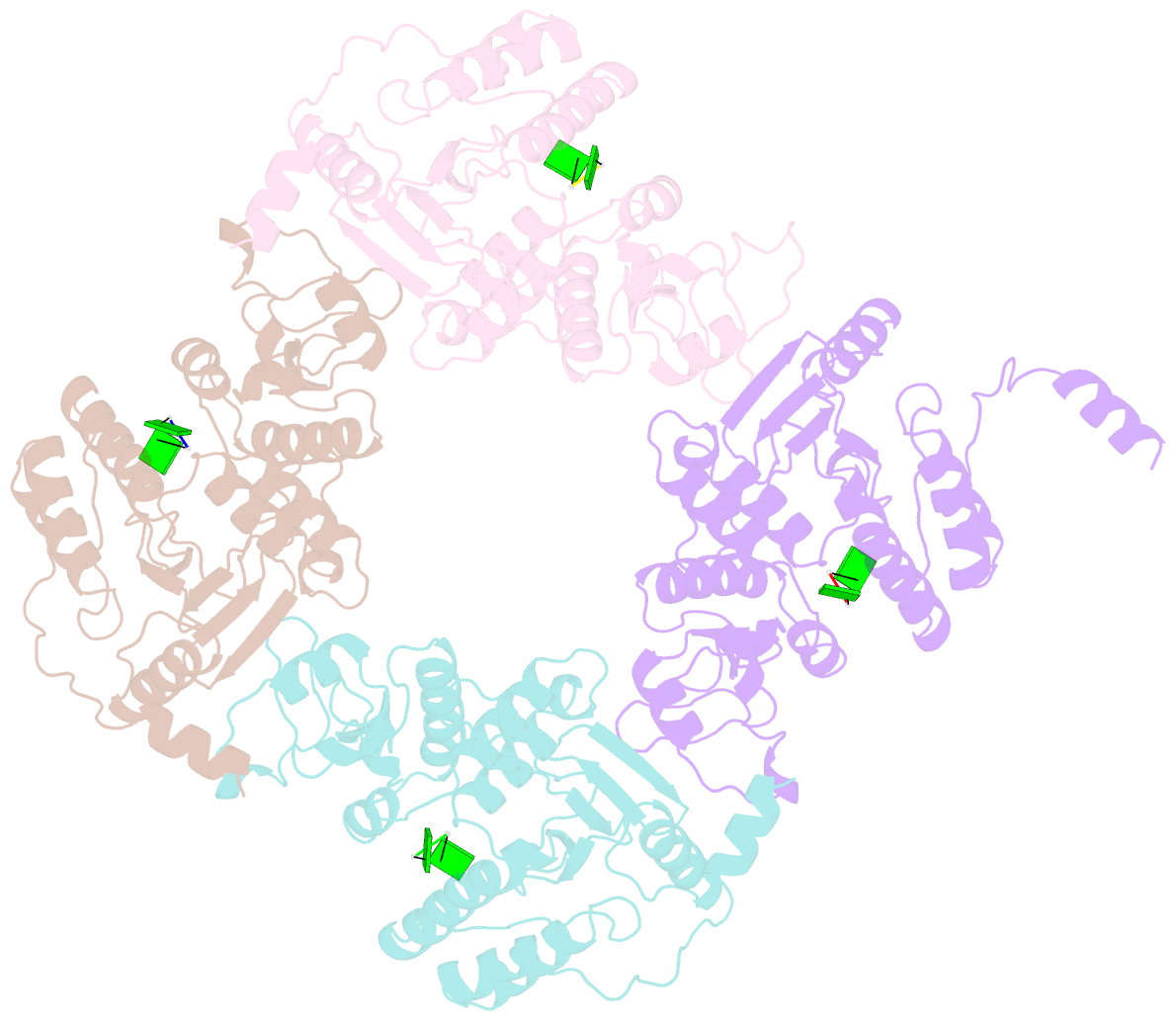
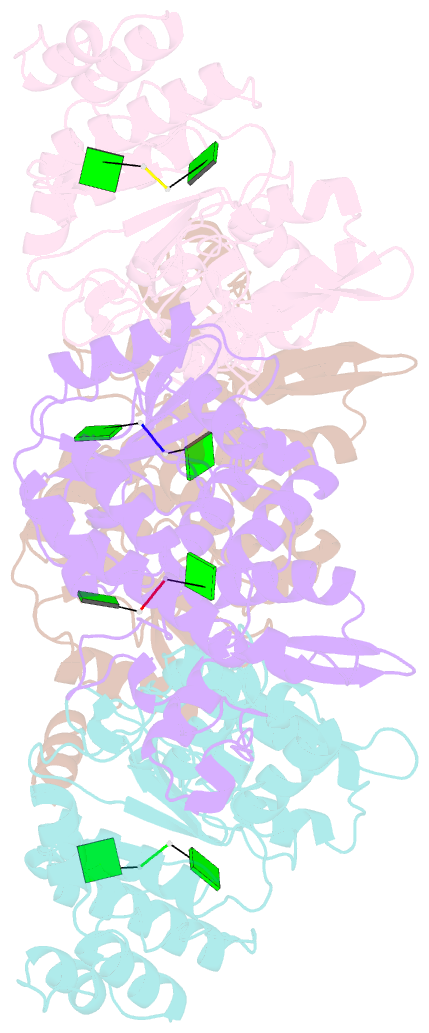
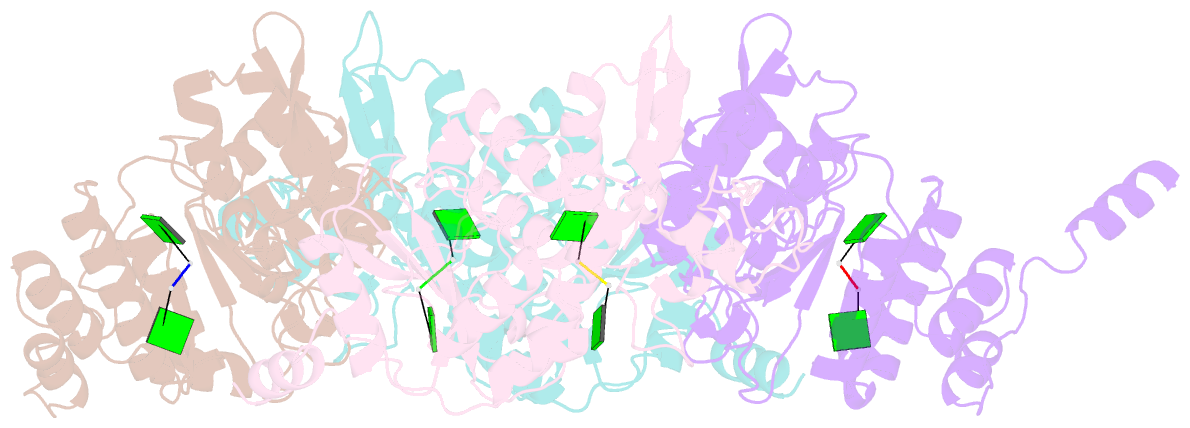
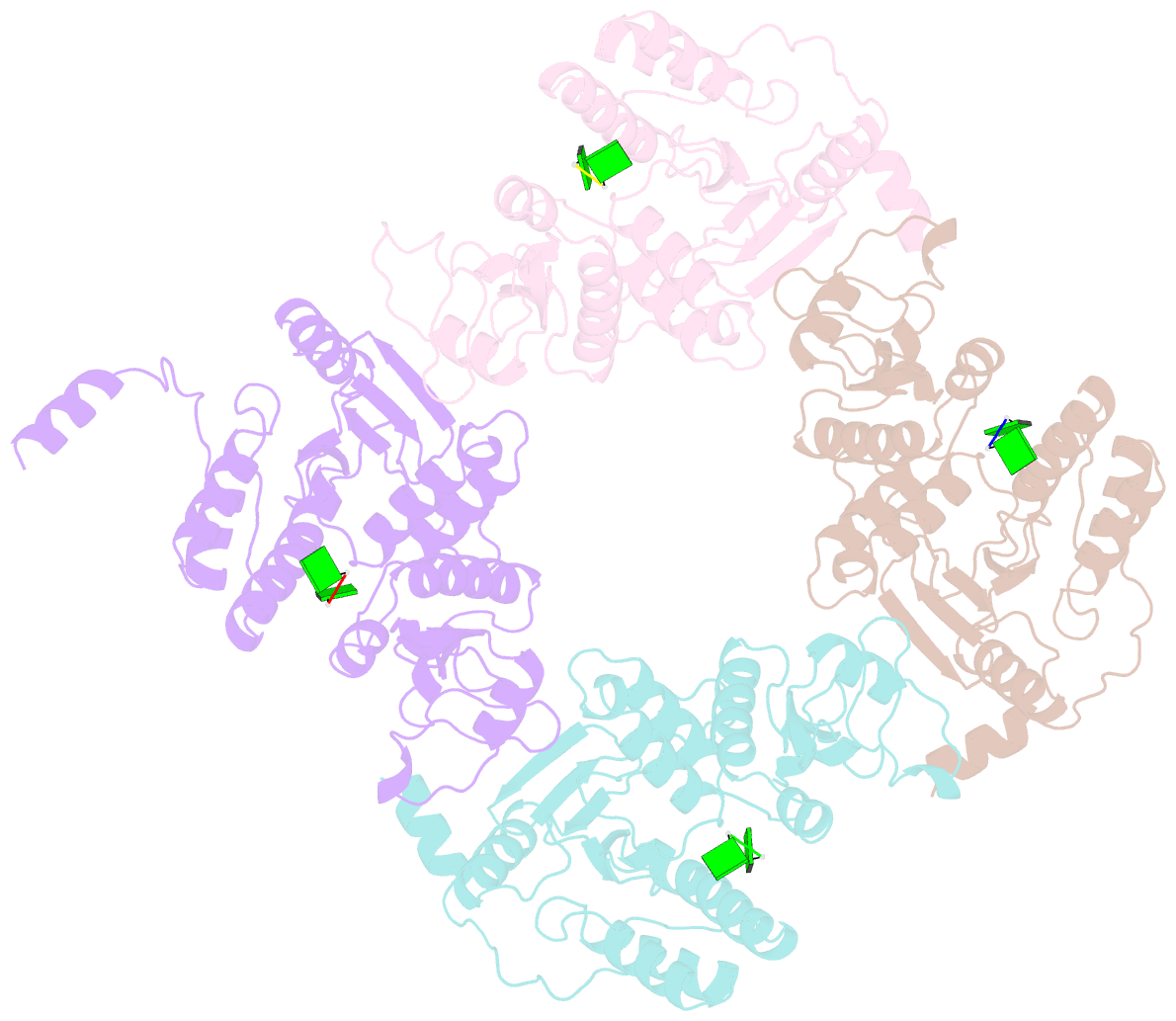
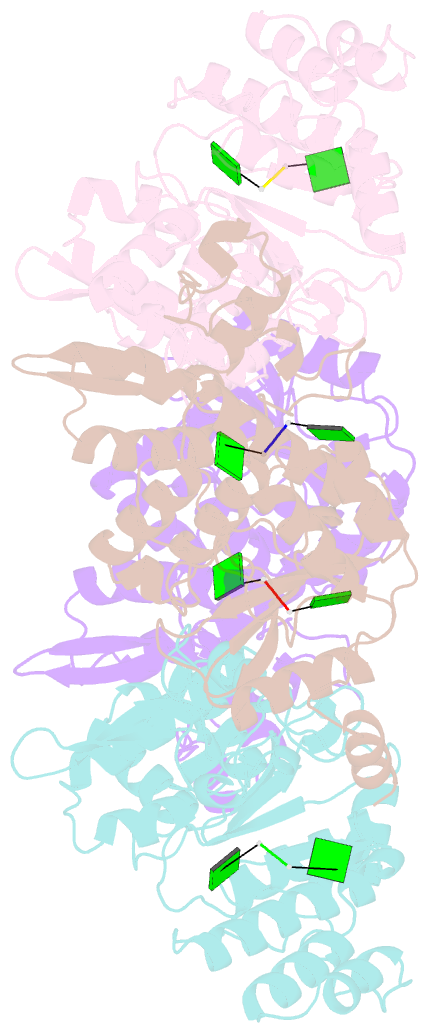
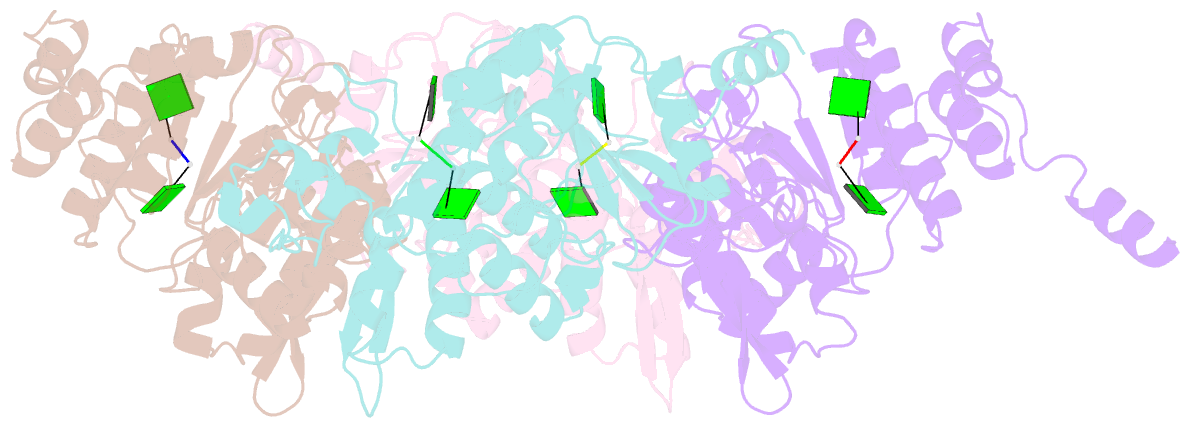
- Crystallographic analysis of rotavirus nsp2-RNA complex reveals specific recognition of 5'-gg sequence for rtpase activity
- Reference
- Hu L, Chow DC, Patton JT, Palzkill T, Estes MK, Prasad BV (2012): "Crystallographic Analysis of Rotavirus NSP2-RNA Complex Reveals Specific Recognition of 5' GG Sequence for RTPase Activity." J.Virol., 86, 10547-10557. doi: 10.1128/JVI.01201-12.
- Abstract
- Rotavirus nonstructural protein NSP2, a functional octamer, is critical for the formation of viroplasms, which are exclusive sites for replication and packaging of the segmented double-stranded RNA (dsRNA) rotavirus genome. As a component of replication intermediates, NSP2 is also implicated in various replication-related activities. In addition to sequence-independent single-stranded RNA-binding and helix-destabilizing activities, NSP2 exhibits monomer-associated nucleoside and 5' RNA triphosphatase (NTPase/RTPase) activities that are mediated by a conserved H225 residue within a narrow enzymatic cleft. Lack of a 5' γ-phosphate is a common feature of the negative-strand RNA [(-)RNA] of the packaged dsRNA segments in rotavirus. Strikingly, all (-)RNAs (of group A rotaviruses) have a 5' GG dinucleotide sequence. As the only rotavirus protein with 5' RTPase activity, NSP2 is implicated in the removal of the γ-phosphate from the rotavirus (-)RNA. To understand how NSP2, despite its sequence-independent RNA-binding property, recognizes (-)RNA to hydrolyze the γ-phosphate within the catalytic cleft, we determined a crystal structure of NSP2 in complex with the 5' consensus sequence of minus-strand rotavirus RNA. Our studies show that the 5' GG of the bound oligoribonucleotide interacts extensively with highly conserved residues in the NSP2 enzymatic cleft. Although these residues provide GG-specific interactions, surface plasmon resonance studies suggest that the C-terminal helix and other basic residues outside the enzymatic cleft account for sequence-independent RNA binding of NSP2. A novel observation from our studies, which may have implications in viroplasm formation, is that the C-terminal helix of NSP2 exhibits two distinct conformations and engages in domain-swapping interactions, which result in the formation of NSP2 octamer chains.