Summary information and primary citation
- PDB-id
- 4gcl; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.65 Å)
- Summary
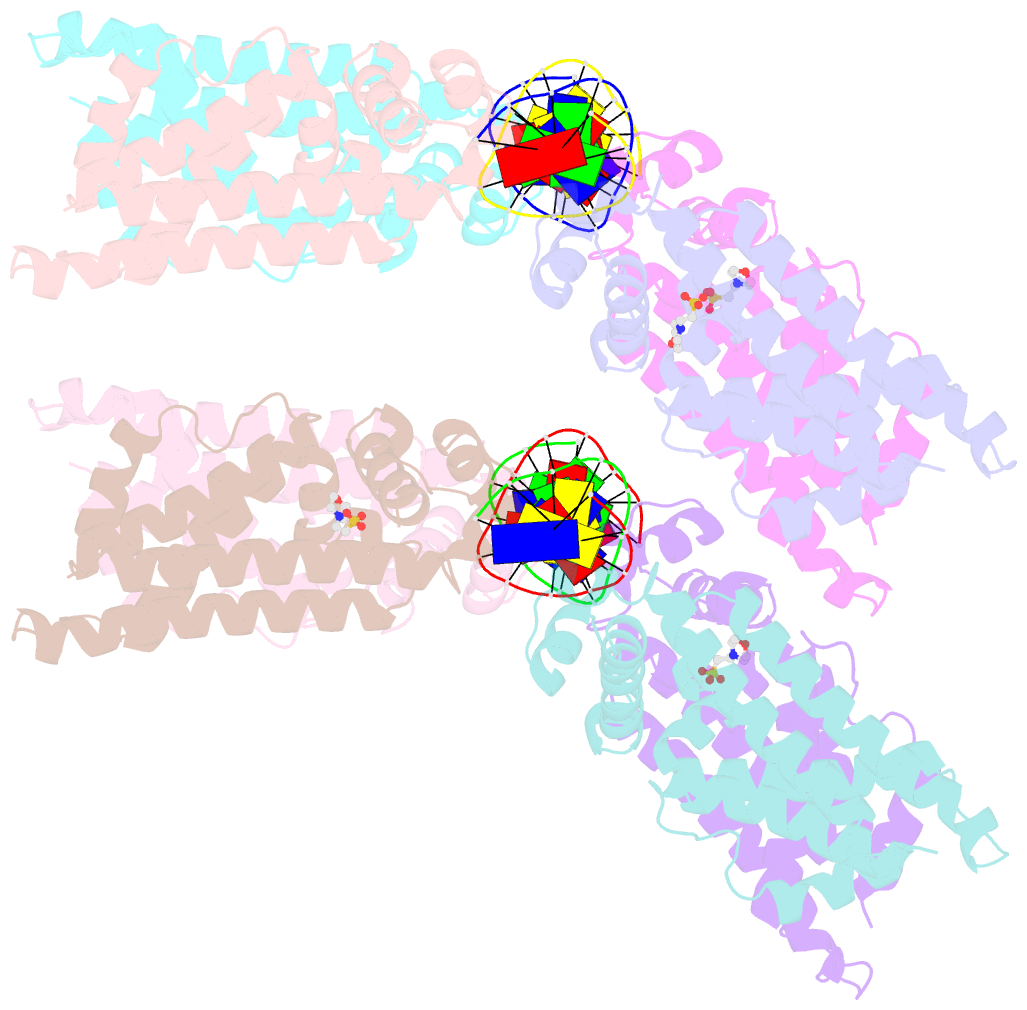
- Structure of no-DNA factor
- Reference
- Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA (2013): "SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid." Proc.Natl.Acad.Sci.USA, 110, 10586-10591. doi: 10.1073/pnas.1221036110.
- Abstract
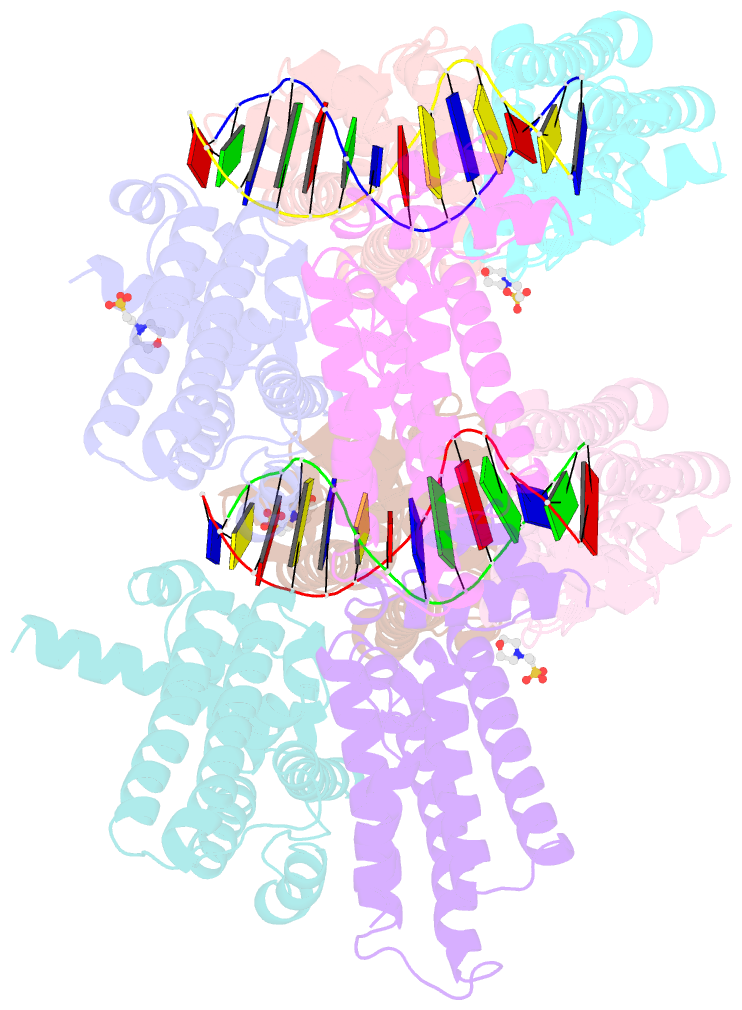
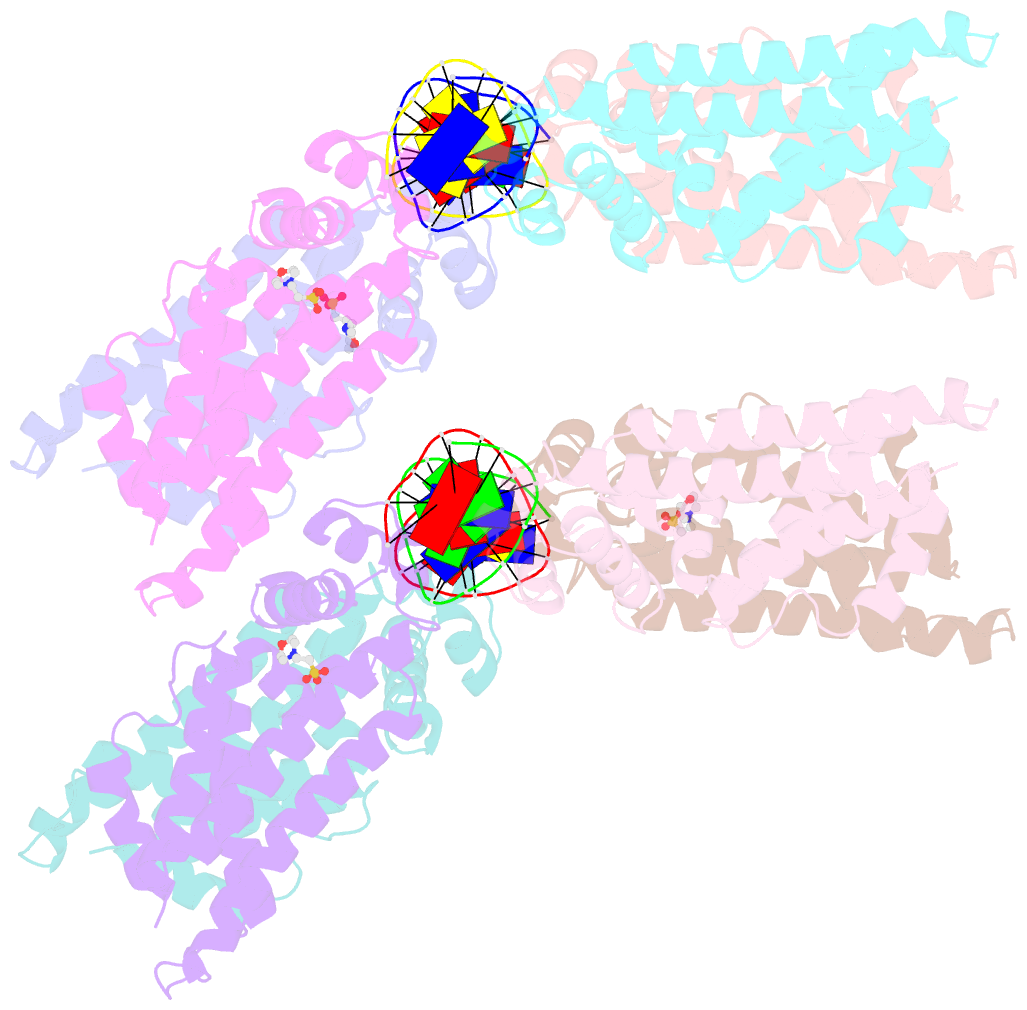
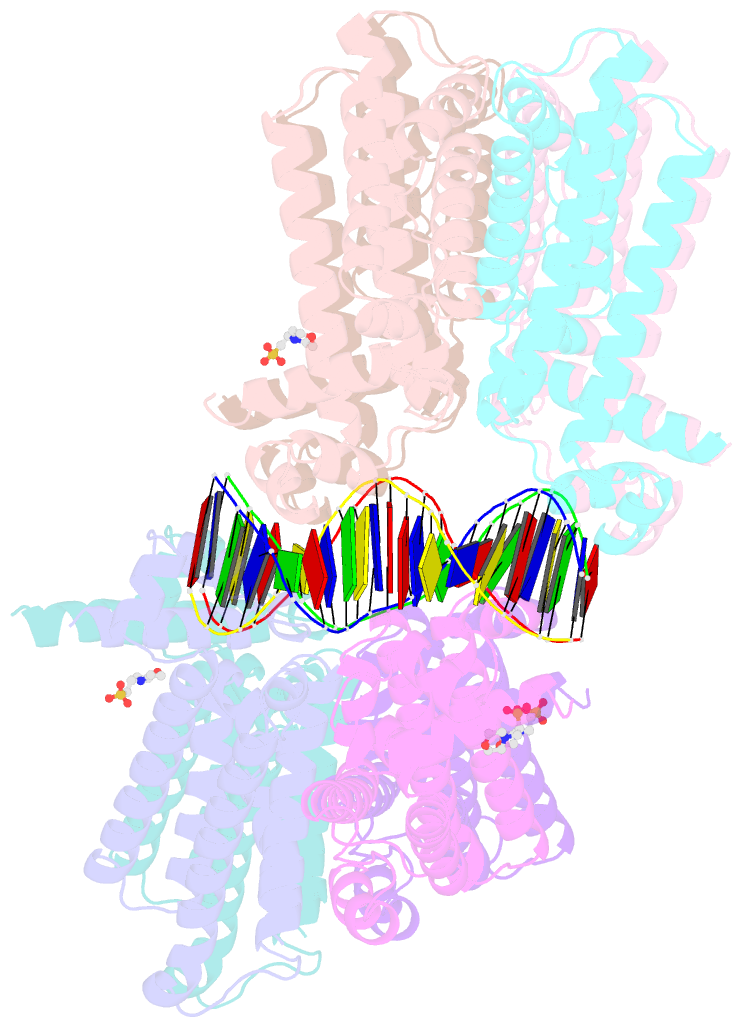
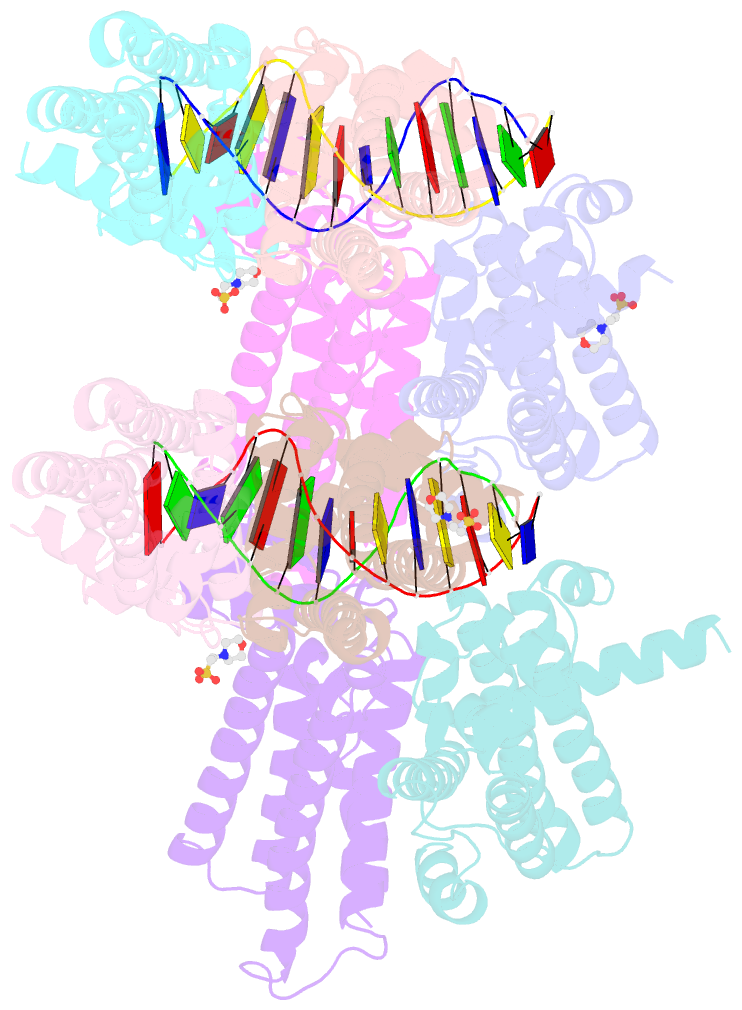
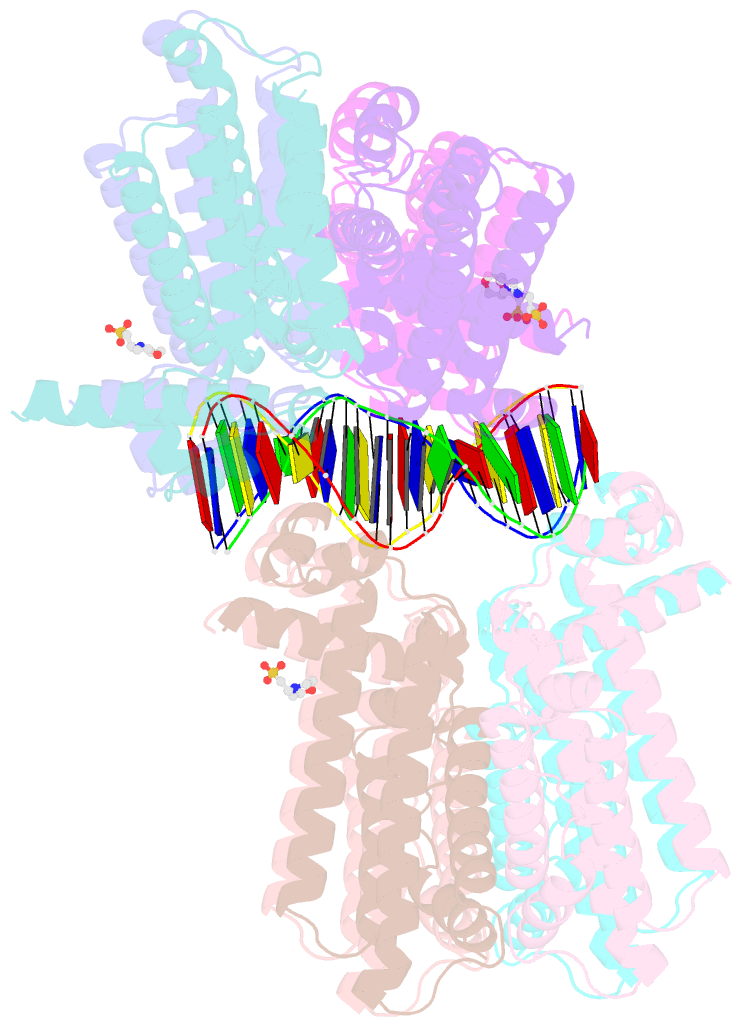
- The spatial and temporal control of Filamenting temperature sensitive mutant Z (FtsZ) Z-ring formation is crucial for proper cell division in bacteria. In Escherichia coli, the synthetic lethal with a defective Min system (SlmA) protein helps mediate nucleoid occlusion, which prevents chromosome fragmentation by binding FtsZ and inhibiting Z-ring formation over the nucleoid. However, to perform its function, SlmA must be bound to the nucleoid. To deduce the basis for this chromosomal requirement, we performed biochemical, cellular, and structural studies. Strikingly, structures show that SlmA dramatically distorts DNA, allowing it to bind as an orientated dimer-of-dimers. Biochemical data indicate that SlmA dimer-of-dimers can spread along the DNA. Combined structural and biochemical data suggest that this DNA-activated SlmA oligomerization would prevent FtsZ protofilament propagation and bundling. Bioinformatic analyses localize SlmA DNA sites near membrane-tethered chromosomal regions, and cellular studies show that SlmA inhibits FtsZ reservoirs from forming membrane-tethered Z rings. Thus, our combined data indicate that SlmA DNA helps block Z-ring formation over chromosomal DNA by forming higher-order protein-nucleic acid complexes that disable FtsZ filaments from coalescing into proper structures needed for Z-ring creation.