Summary information and primary citation
- PDB-id
- 4gfh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA
- Method
- X-ray (4.408 Å)
- Summary
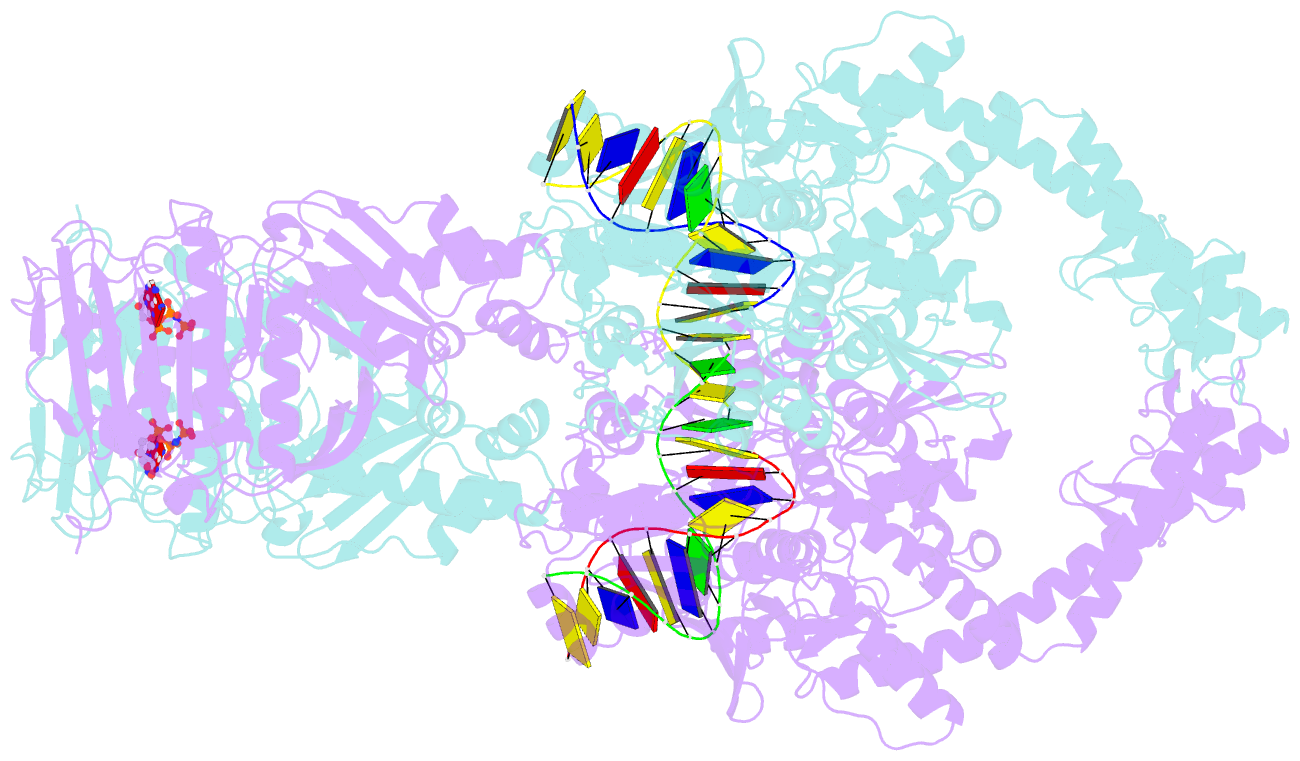
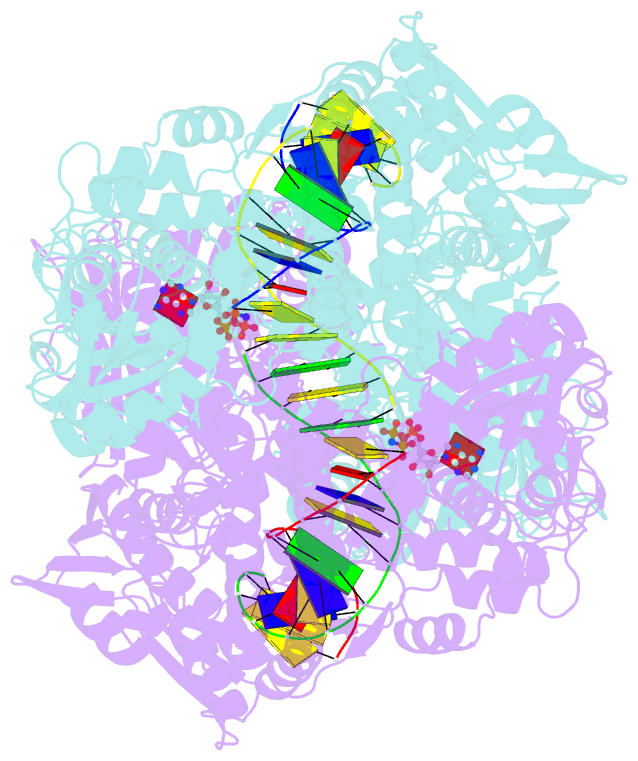
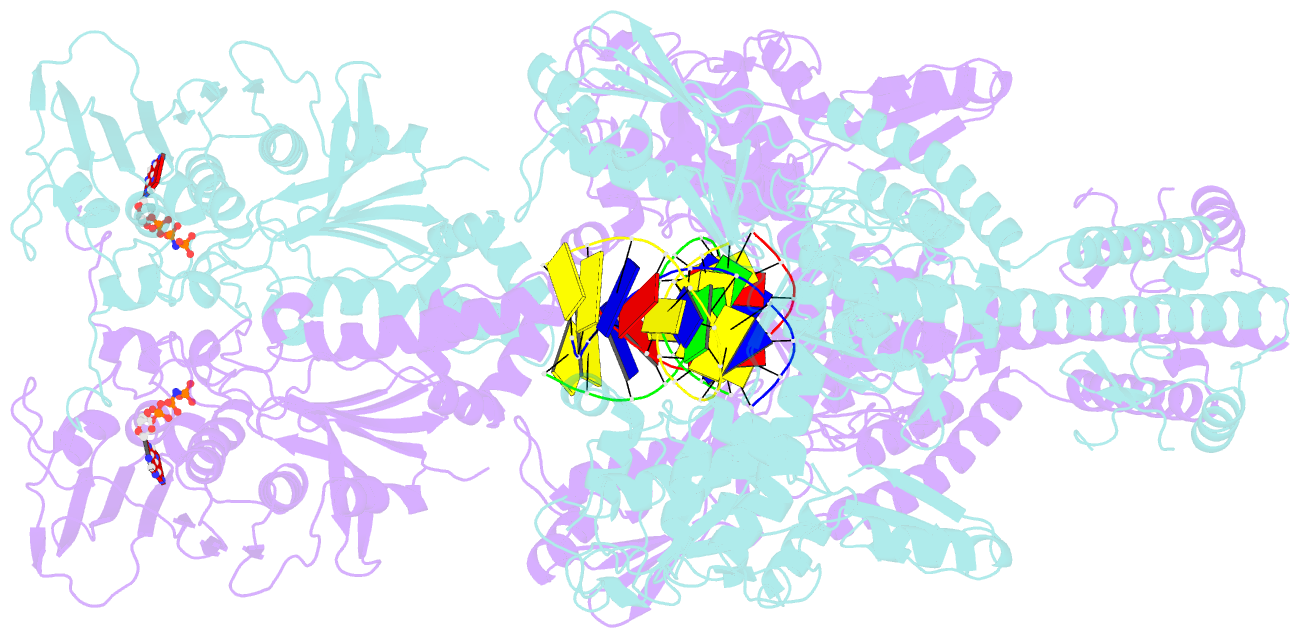
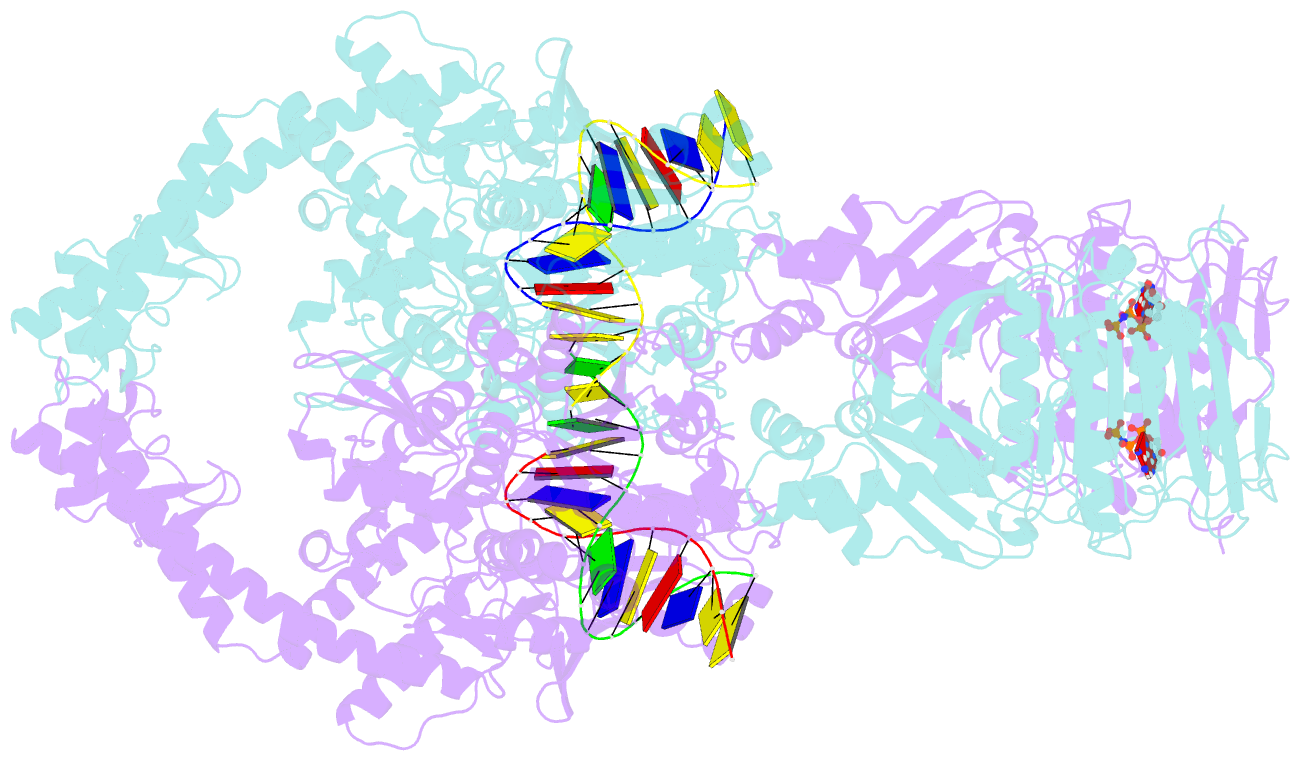
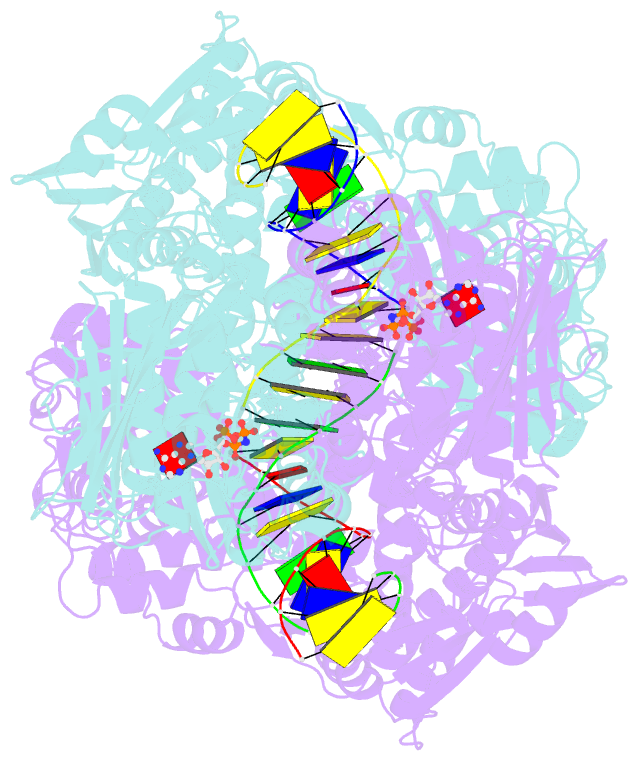
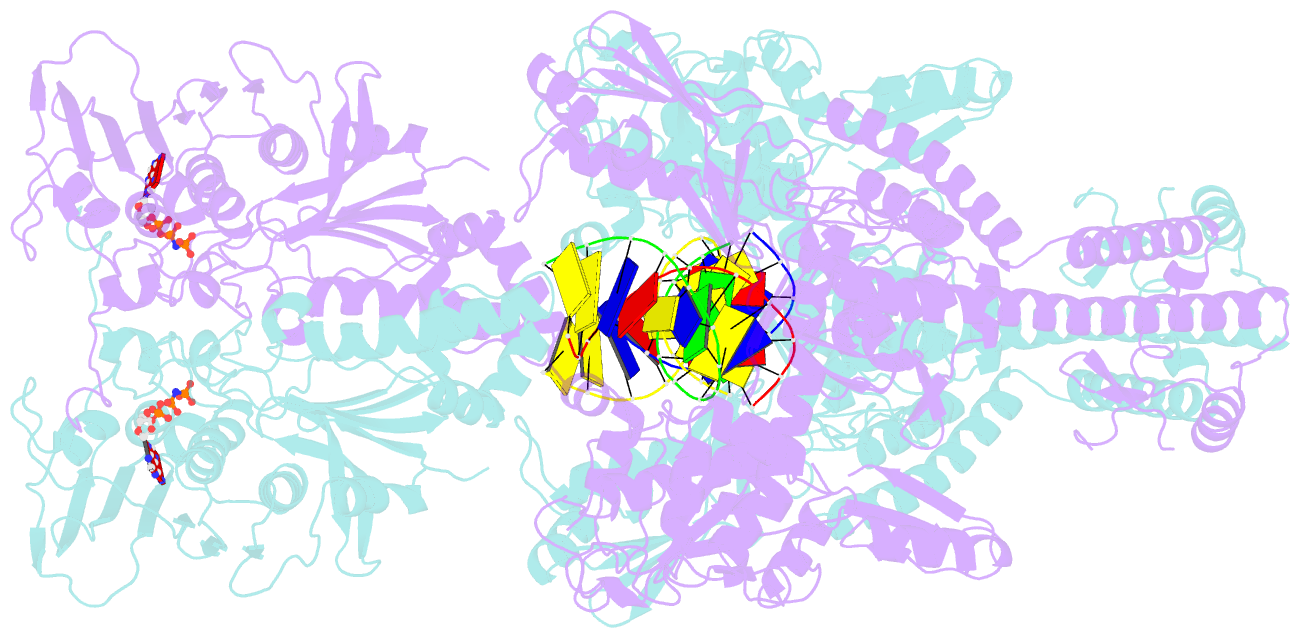
- Topoisomerase ii-DNA-amppnp complex
- Reference
- Schmidt BH, Osheroff N, Berger JM (2012): "Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity." Nat.Struct.Mol.Biol., 19, 1147-1154. doi: 10.1038/nsmb.2388.
- Abstract
- Type IIA topoisomerases control DNA supercoiling and disentangle chromosomes through a complex ATP-dependent strand-passage mechanism. Although a general framework exists for type IIA topoisomerase function, the architecture of the full-length enzyme has remained undefined. Here we present the structure of a fully catalytic Saccharomyces cerevisiae topoisomerase II homodimer complexed with DNA and a nonhydrolyzable ATP analog. The enzyme adopts a domain-swapped configuration wherein the ATPase domain of one protomer sits atop the nucleolytic region of its partner subunit. This organization produces an unexpected interaction between bound DNA and a conformational transducing element in the ATPase domain, which we show is critical for both DNA-stimulated ATP hydrolysis and global topoisomerase activity. Our data indicate that the ATPase domains pivot about each other to ensure unidirectional strand passage and that this state senses bound DNA to promote ATP turnover and enzyme reset.