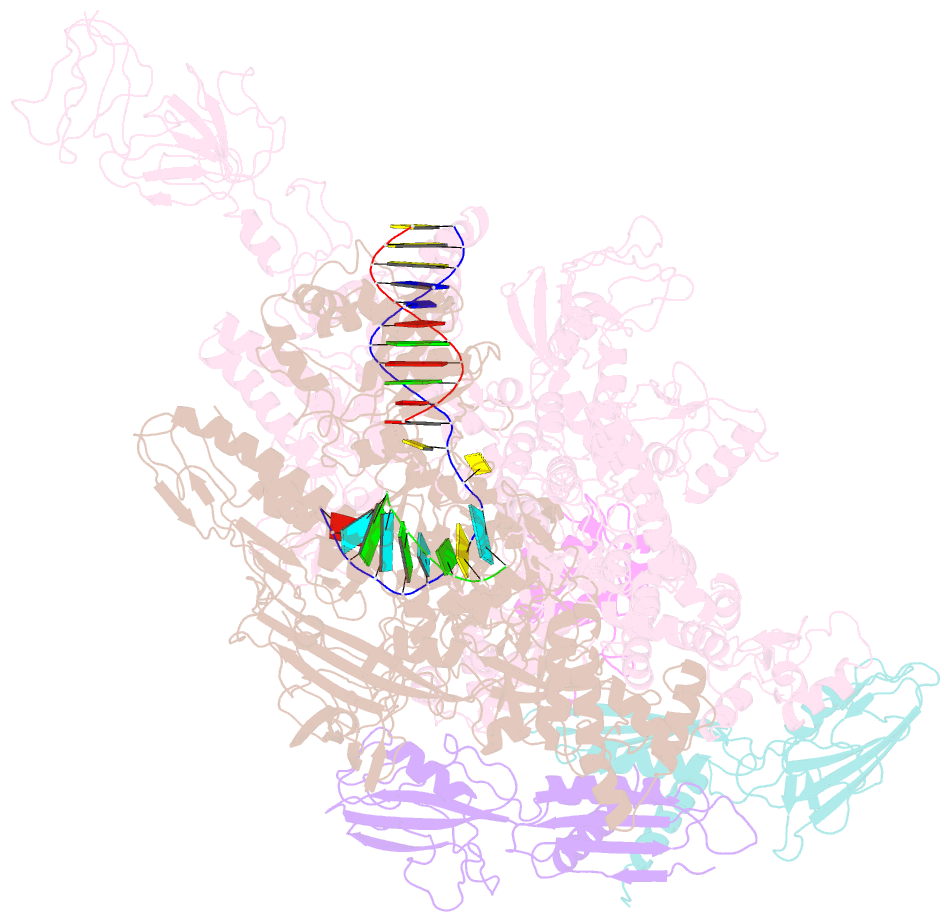
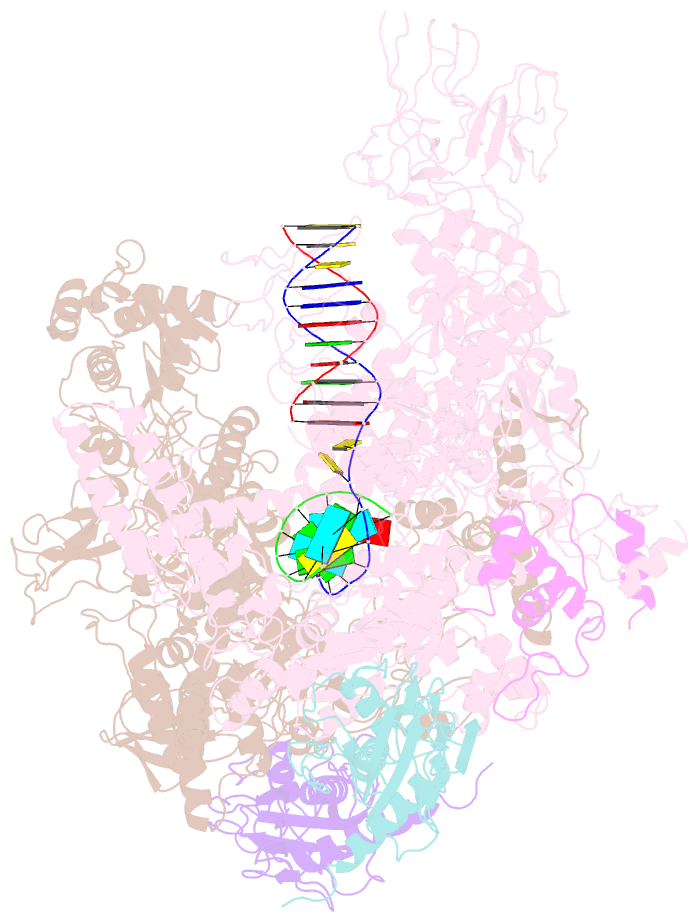
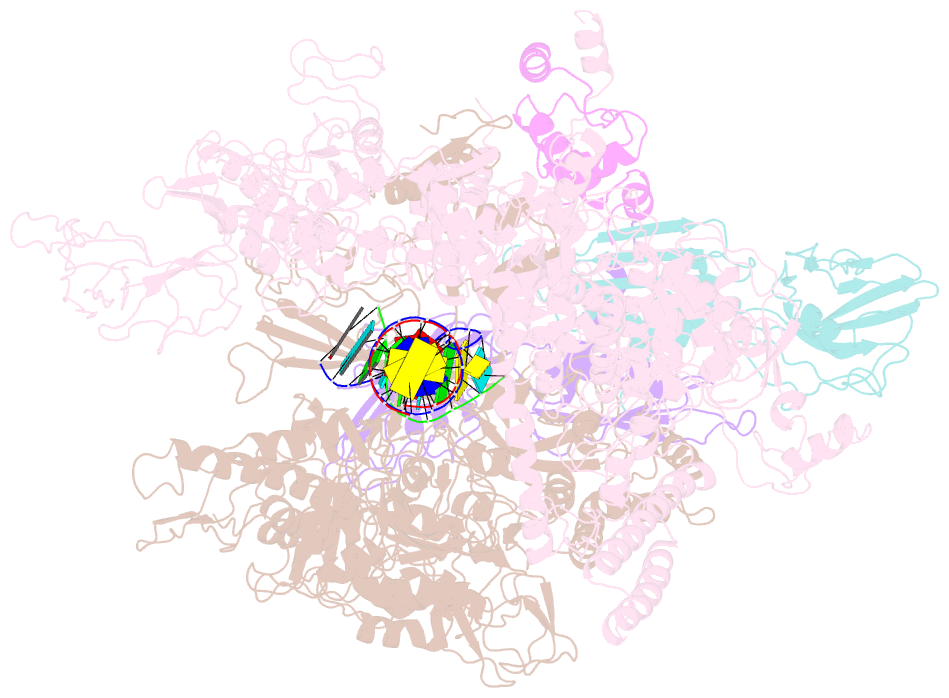
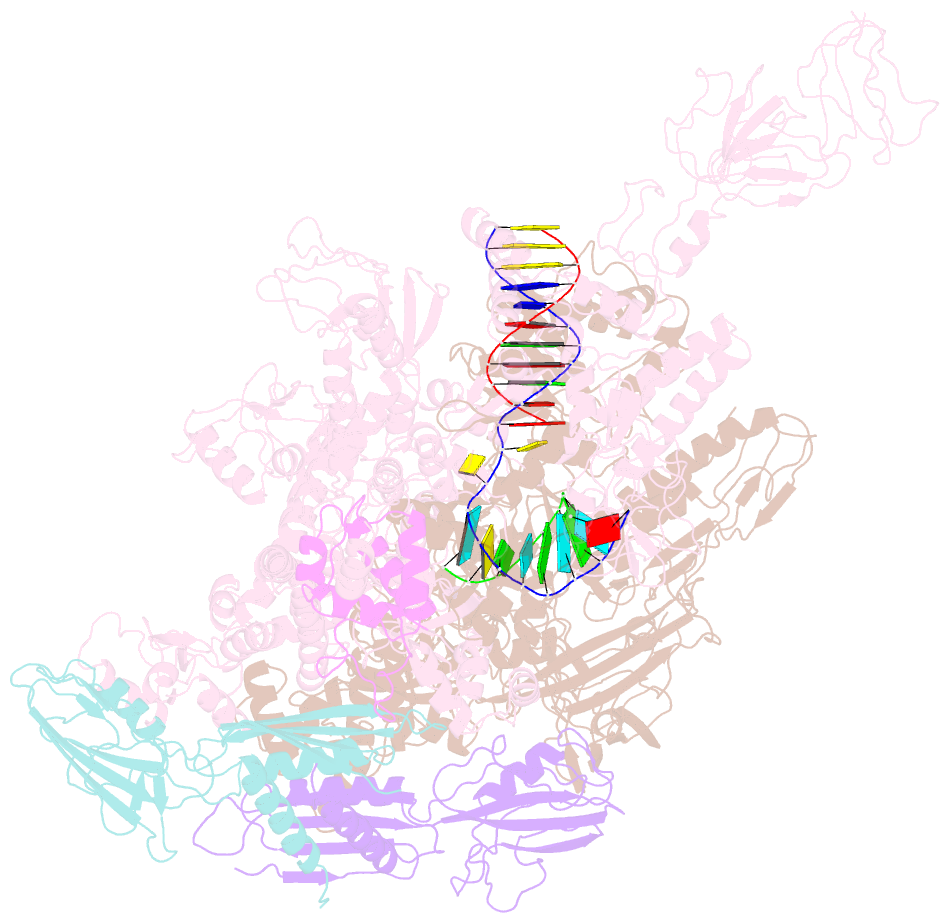
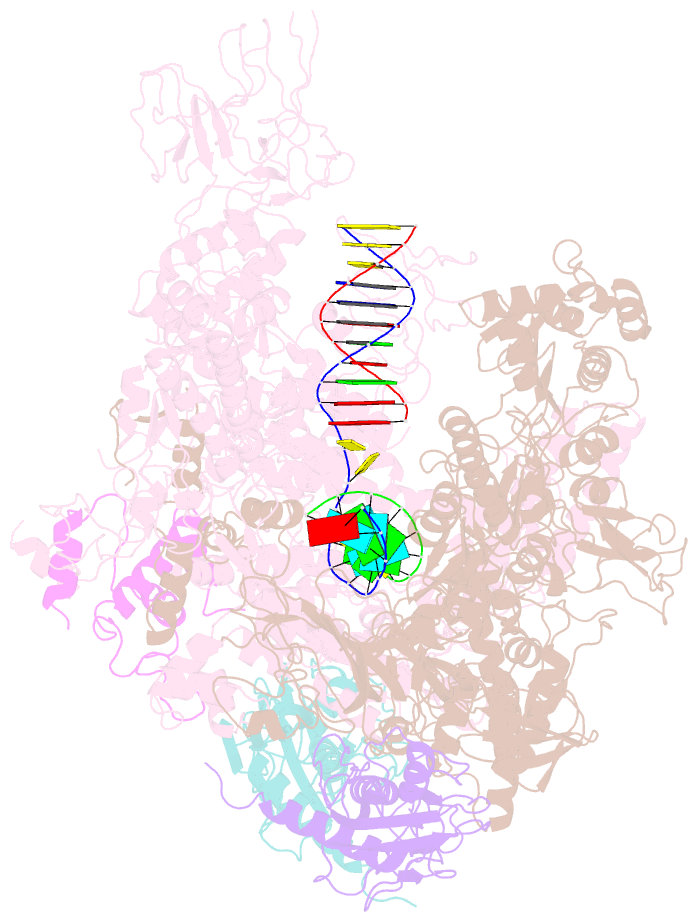
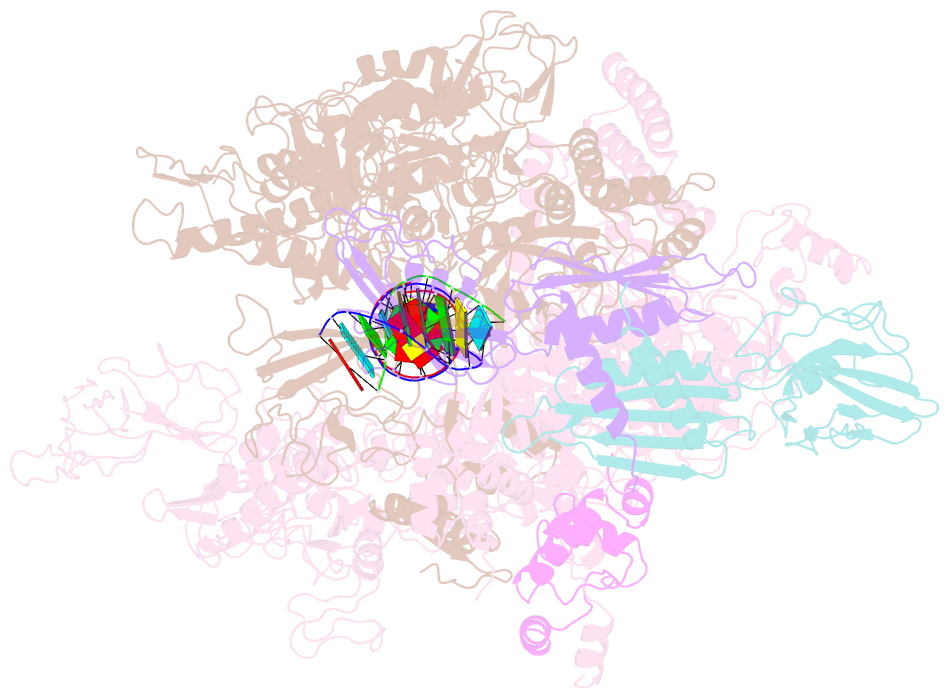
Summary information and primary citation
- PDB-id
- 4gzz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA-RNA hybrid
- Method
- X-ray (4.29 Å)
- Summary
- Crystal structures of bacterial RNA polymerase paused elongation complexes
- Reference
- Weixlbaumer A, Leon K, Landick R, Darst SA (2013): "Structural basis of transcriptional pausing in bacteria." Cell(Cambridge,Mass.), 152, 431-441. doi: 10.1016/j.cell.2012.12.020.
- Abstract
- Transcriptional pausing by multisubunit RNA polymerases (RNAPs) is a key mechanism for regulating gene expression in both prokaryotes and eukaryotes and is a prerequisite for transcription termination. Pausing and termination states are thought to arise through a common, elemental pause state that is inhibitory for nucleotide addition. We report three crystal structures of Thermus RNAP elemental paused elongation complexes (ePECs). The structures reveal the same relaxed, open-clamp RNAP conformation in the ePEC that may arise by failure to re-establish DNA contacts during translocation. A kinked bridge-helix sterically blocks the RNAP active site, explaining how this conformation inhibits RNAP catalytic activity. Our results provide a framework for understanding how RNA hairpin formation stabilizes the paused state and how the ePEC intermediate facilitates termination.