Summary information and primary citation
- PDB-id
- 4hc7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.65 Å)
- Summary
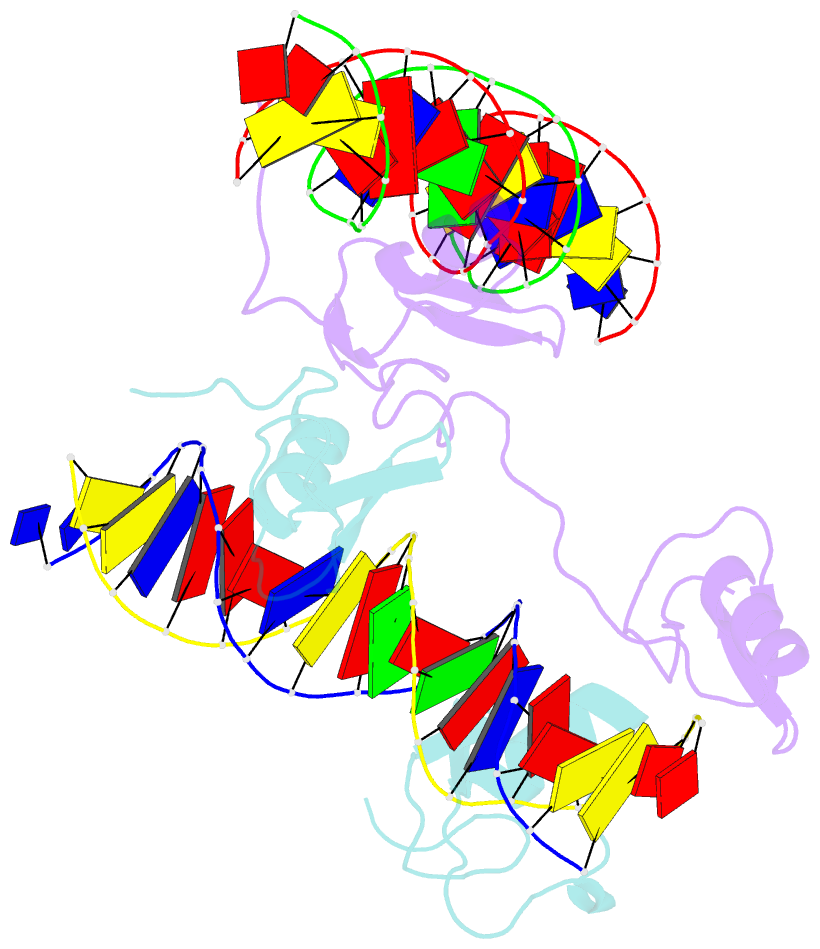
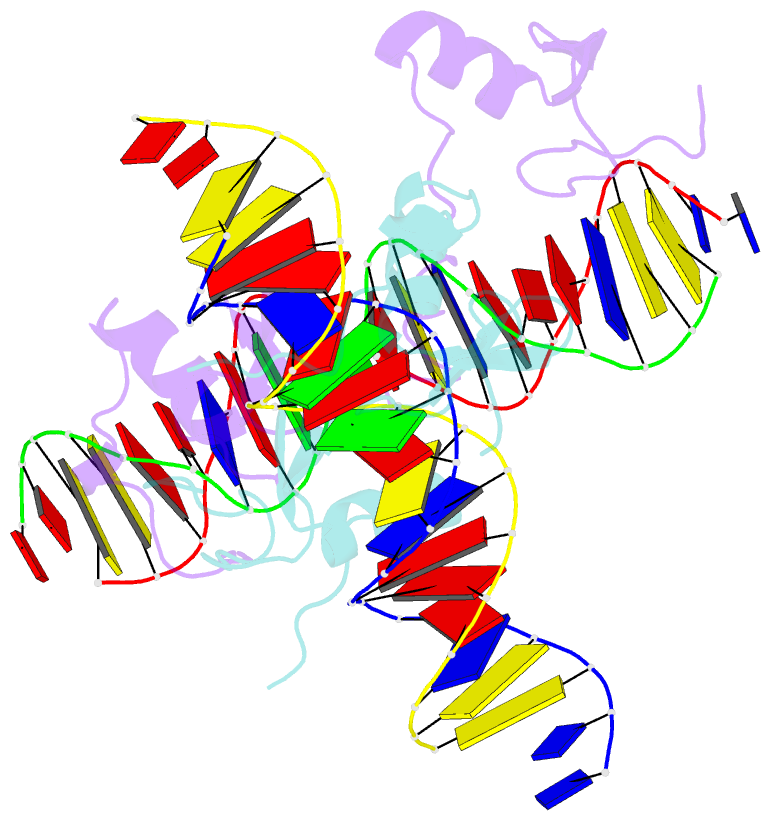
- Crystal structure of the full DNA binding domain of gata3-complex 2
- Reference
- Chen Y, Bates DL, Dey R, Chen PH, Machado AC, Laird-Offringa IA, Rohs R, Chen L (2012): "DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation." Cell Rep, 2, 1197-1206. doi: 10.1016/j.celrep.2012.10.012.
- Abstract
- GATA transcription factors regulate transcription during development and differentiation by recognizing distinct GATA sites with a tandem of two conserved zinc fingers, and by mediating long-range DNA looping. However, the molecular basis of these processes is not well understood. Here, we determined three crystal structures of the full DNA-binding domain (DBD) of human GATA3 protein, which contains both zinc fingers, in complex with different DNA sites. In one structure, both zinc fingers wrap around a palindromic GATA site, cooperatively enhancing the binding affinity and kinetic stability. Strikingly, in the other two structures, the two fingers of GATA DBD bind GATA sites on different DNA molecules, thereby bridging two separate DNA fragments. This was confirmed in solution by an in-gel fluorescence resonance energy transfer analysis. These findings not only provide insights into the structure and function of GATA proteins but also shed light on the molecular basis of long-range gene regulation.