Summary information and primary citation
- PDB-id
- 4hiv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA-antibiotic
- Method
- X-ray (2.6 Å)
- Summary
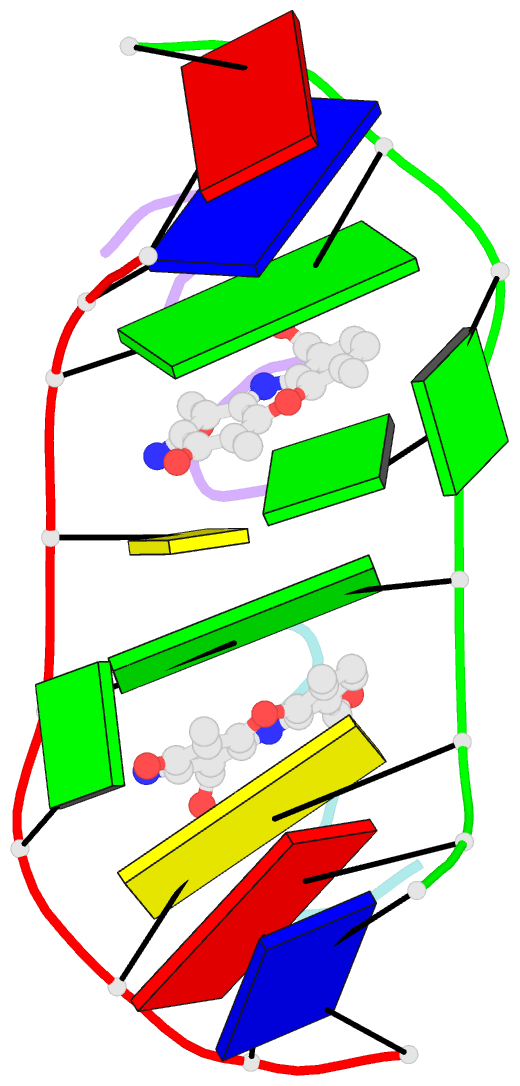
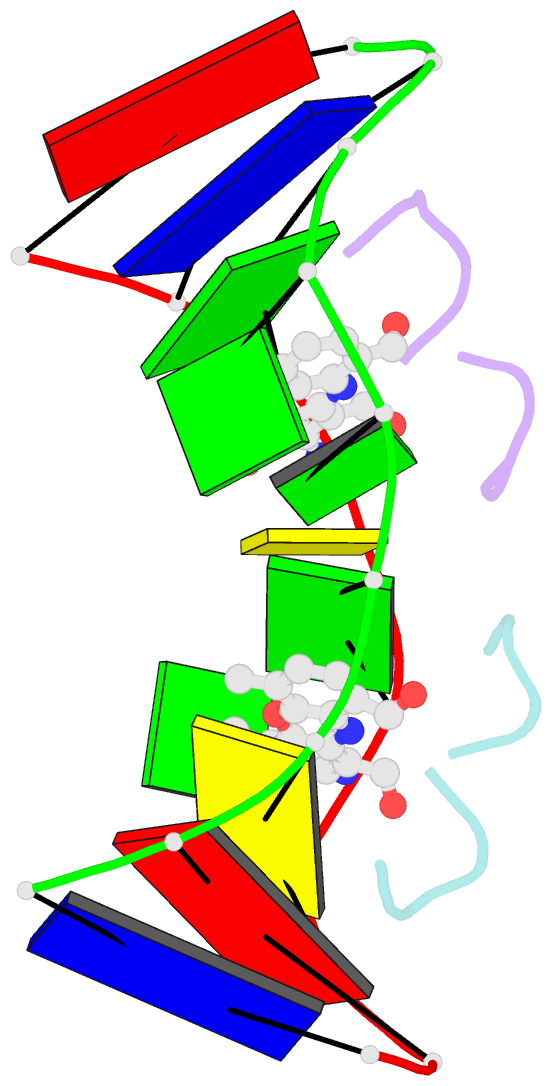
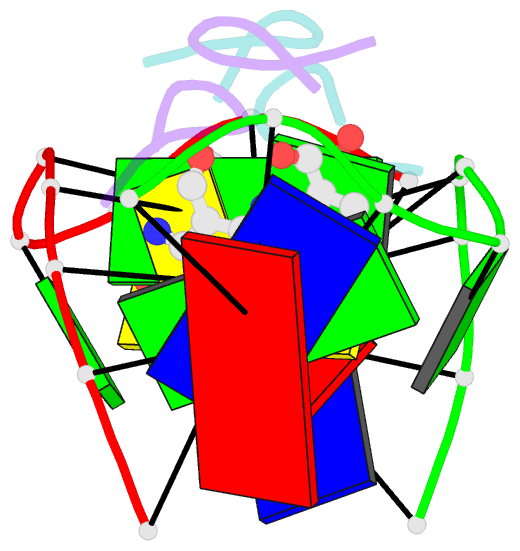
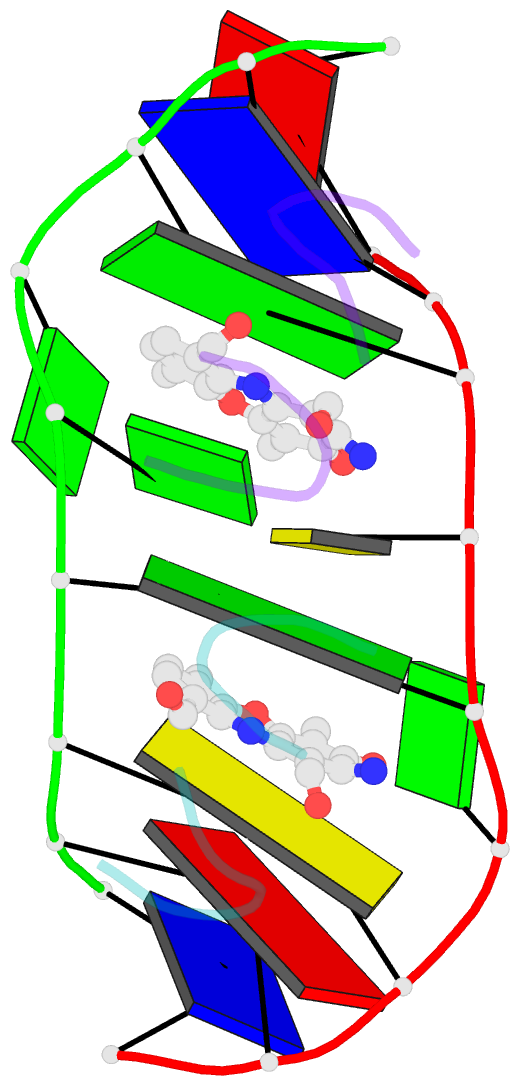
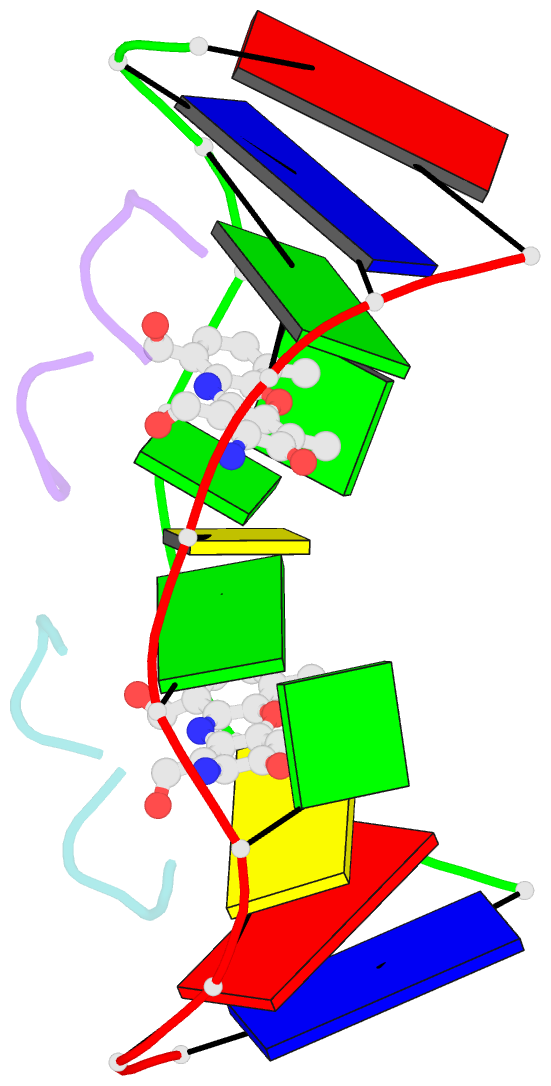
- Structure of actinomycin d d(atgcggcat) complex
- Reference
- Lo YS, Tseng WH, Chuang CY, Hou MH (2013): "The structural basis of actinomycin D-binding induces nucleotide flipping out, a sharp bend and a left-handed twist in CGG triplet repeats." Nucleic Acids Res., 41, 4284-4294. doi: 10.1093/nar/gkt084.
- Abstract
- The potent anticancer drug actinomycin D (ActD) functions by intercalating into DNA at GpC sites, thereby interrupting essential biological processes including replication and transcription. Certain neurological diseases are correlated with the expansion of (CGG)n trinucleotide sequences, which contain many contiguous GpC sites separated by a single G:G mispair. To characterize the binding of ActD to CGG triplet repeat sequences, the structural basis for the strong binding of ActD to neighbouring GpC sites flanking a G:G mismatch has been determined based on the crystal structure of ActD bound to ATGCGGCAT, which contains a CGG triplet sequence. The binding of ActD molecules to GCGGC causes many unexpected conformational changes including nucleotide flipping out, a sharp bend and a left-handed twist in the DNA helix via a two site-binding model. Heat denaturation, circular dichroism and surface plasmon resonance analyses showed that adjacent GpC sequences flanking a G:G mismatch are preferred ActD-binding sites. In addition, ActD was shown to bind the hairpin conformation of (CGG)16 in a pairwise combination and with greater stability than that of other DNA intercalators. Our results provide evidence of a possible biological consequence of ActD binding to CGG triplet repeat sequences.