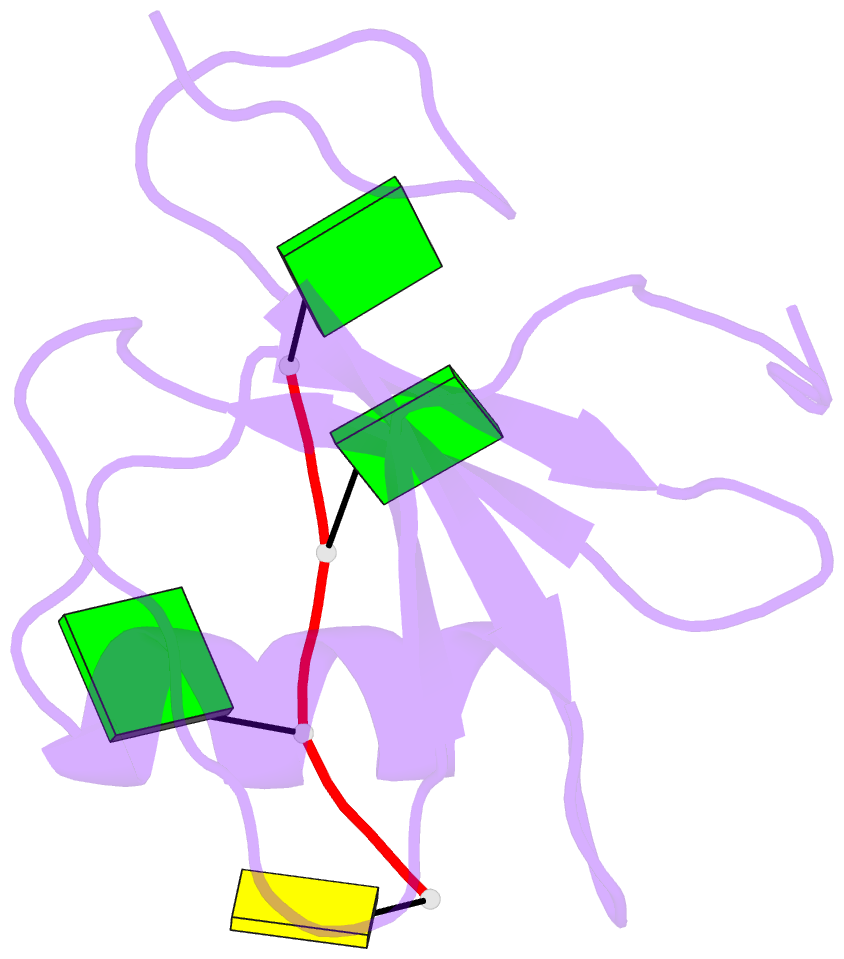
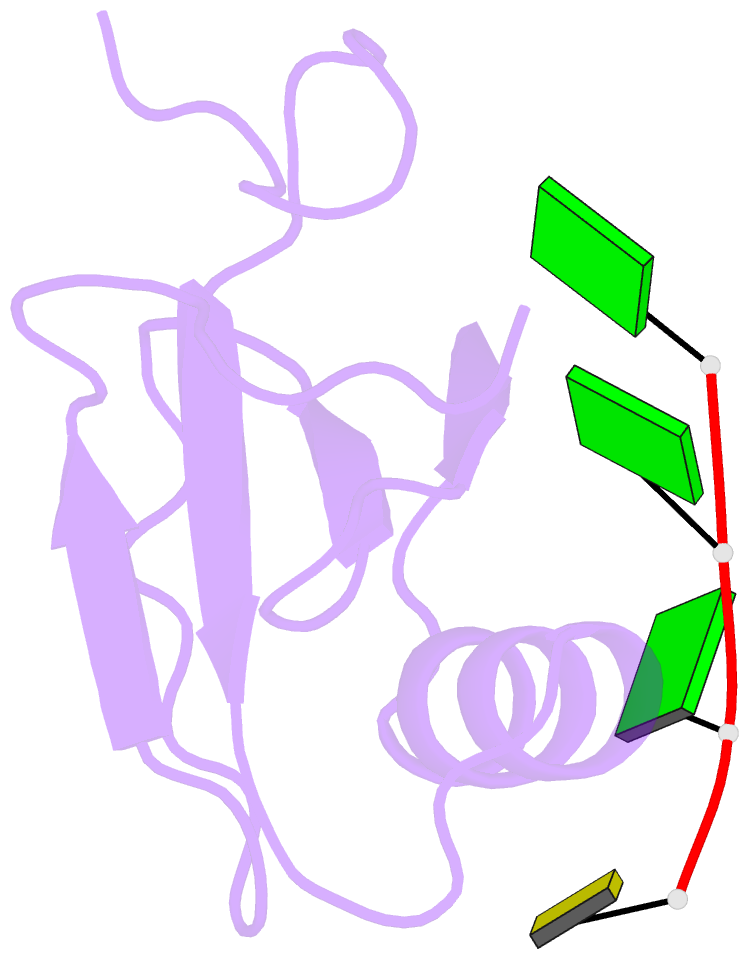
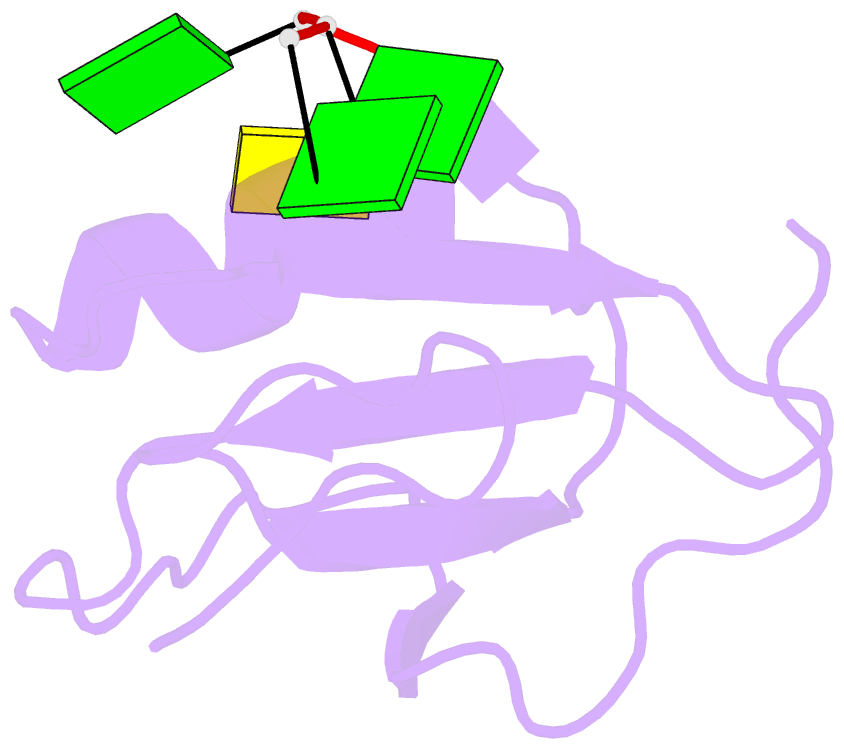
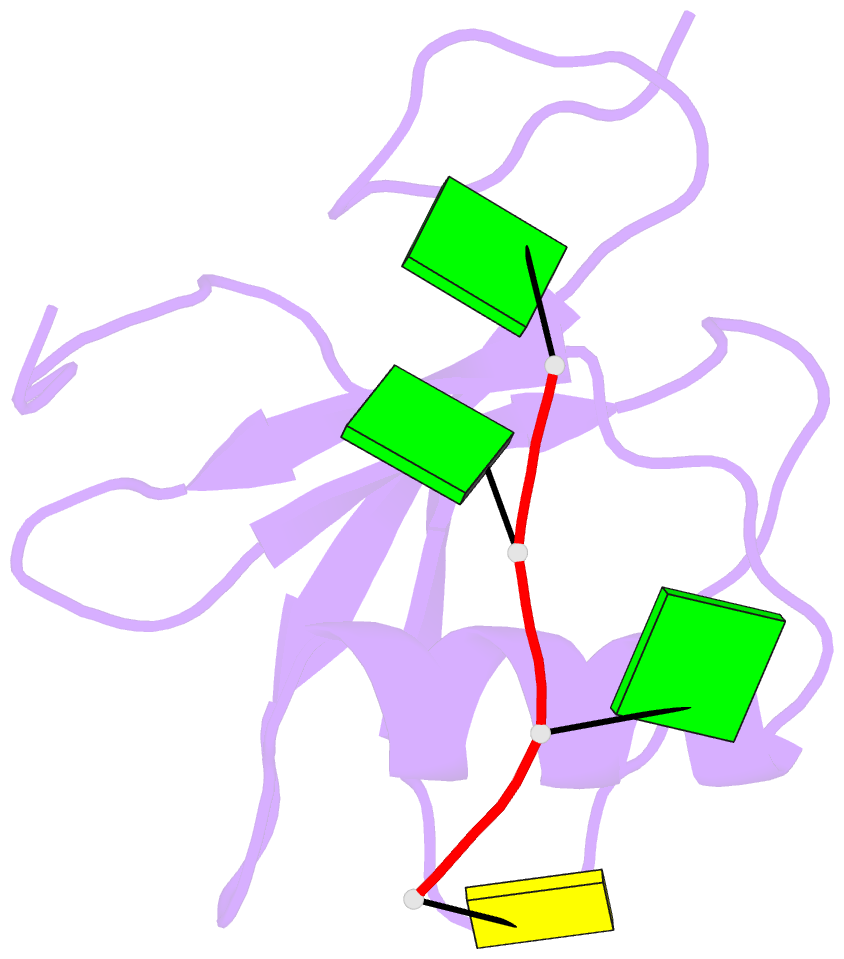
Summary information and primary citation
- PDB-id
- 4i67; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.33 Å)
- Summary
- Crystal structure of the rrm domain of RNA helicase hera from t. thermophilus in complex with gggc RNA
- Reference
- Steimer L, Wurm JP, Linden MH, Rudolph MG, Wohnert J, Klostermeier D (2013): "Recognition of two distinct elements in the RNA substrate by the RNA-binding domain of the T. thermophilus DEAD box helicase Hera." Nucleic Acids Res., 41, 6259-6272. doi: 10.1093/nar/gkt323.
- Abstract
- DEAD box helicases catalyze the ATP-dependent destabilization of RNA duplexes. Whereas duplex separation is mediated by the helicase core shared by all members of the family, flanking domains often contribute to binding of the RNA substrate. The Thermus thermophilus DEAD-box helicase Hera (for "heat-resistant RNA-binding ATPase") contains a C-terminal RNA-binding domain (RBD). We have analyzed RNA binding to the Hera RBD by a combination of mutational analyses, nuclear magnetic resonance and X-ray crystallography, and identify residues on helix α1 and the C-terminus as the main determinants for high-affinity RNA binding. A crystal structure of the RBD in complex with a single-stranded RNA resolves the RNA-protein interactions in the RBD core region around helix α1. Differences in RNA binding to the Hera RBD and to the structurally similar RBD of the Bacillus subtilis DEAD box helicase YxiN illustrate the versatility of RNA recognition motifs as RNA-binding platforms. Comparison of chemical shift perturbation patterns elicited by different RNAs, and the effect of sequence changes in the RNA on binding and unwinding show that the RBD binds a single-stranded RNA region at the core and simultaneously contacts double-stranded RNA through its C-terminal tail. The helicase core then unwinds an adjacent RNA duplex. Overall, the mode of RNA binding by Hera is consistent with a possible function as a general RNA chaperone.