Summary information and primary citation
- PDB-id
- 4i7y; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-hydrolase inhibitor-DNA
- Method
- X-ray (2.4 Å)
- Summary
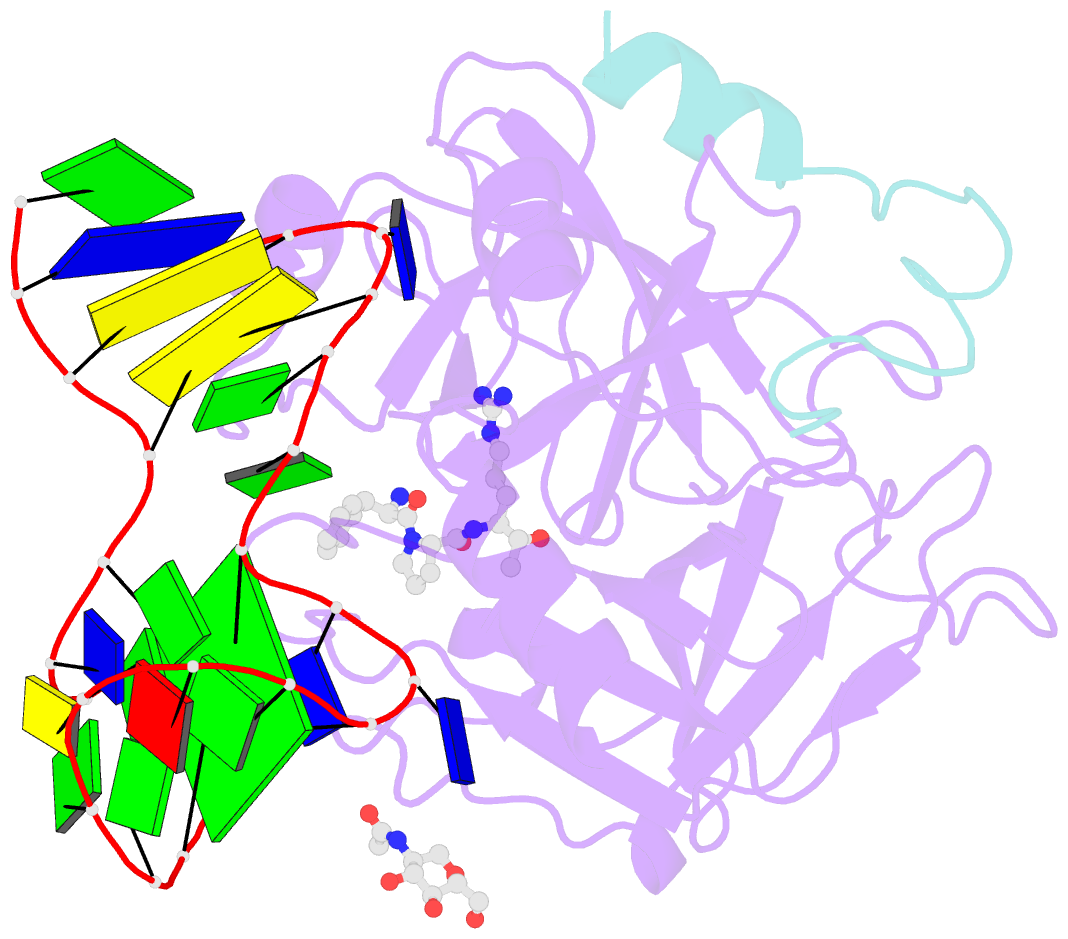
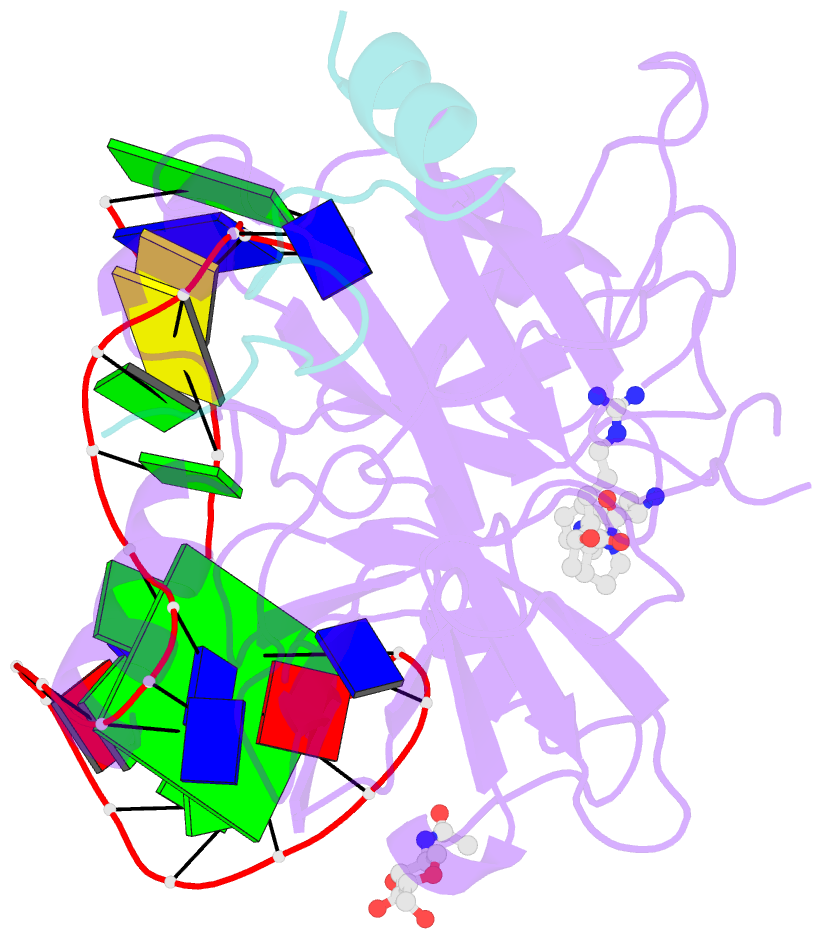
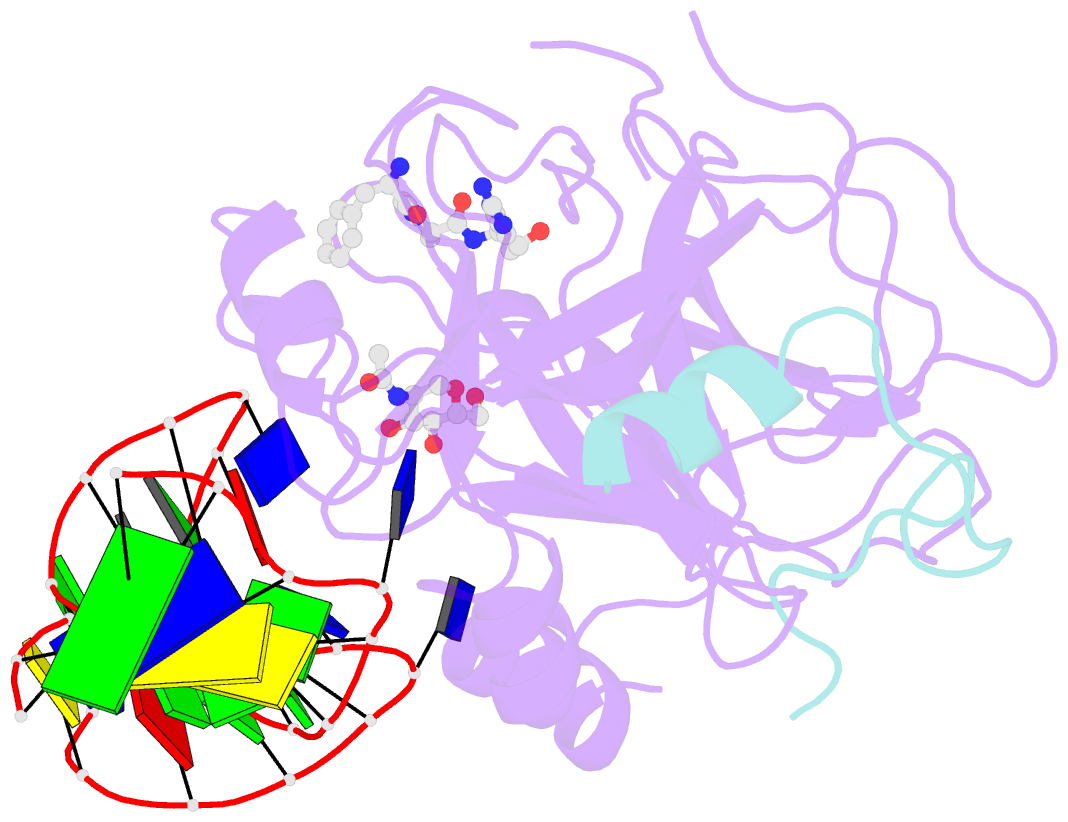
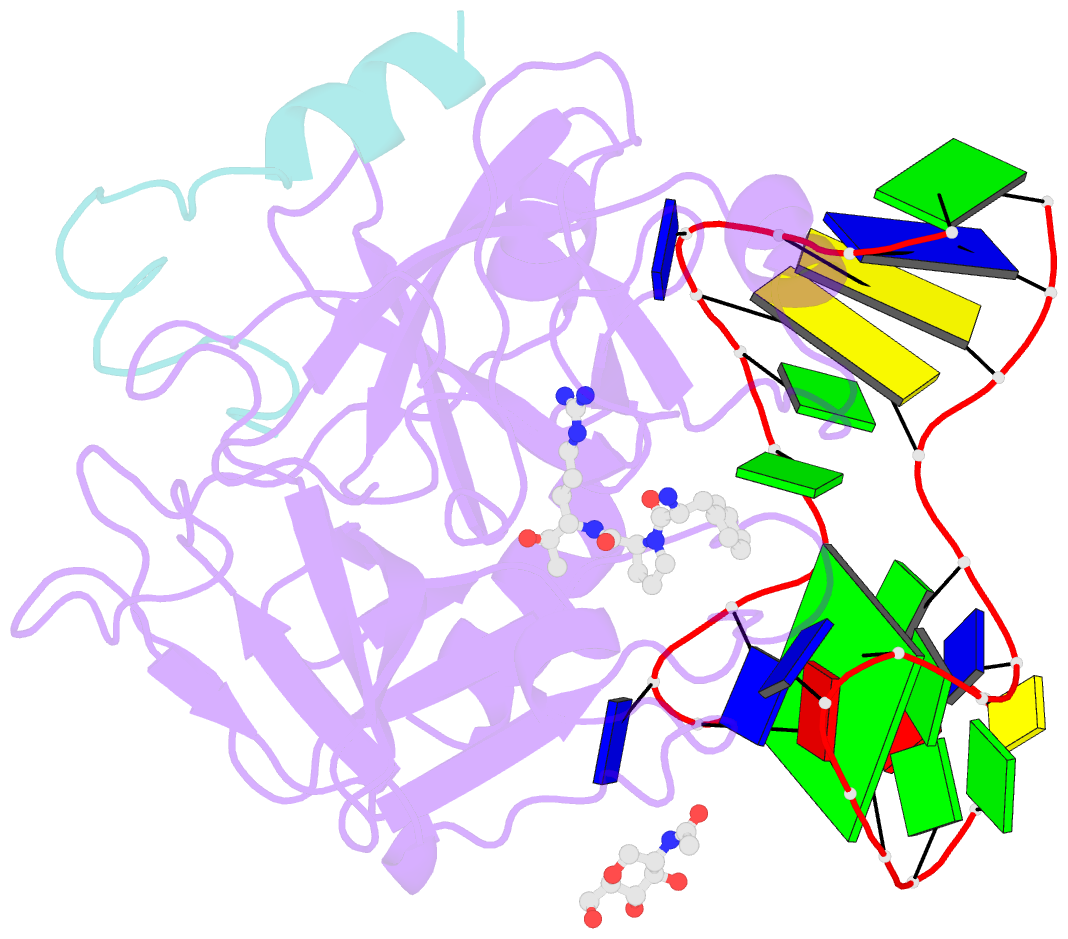
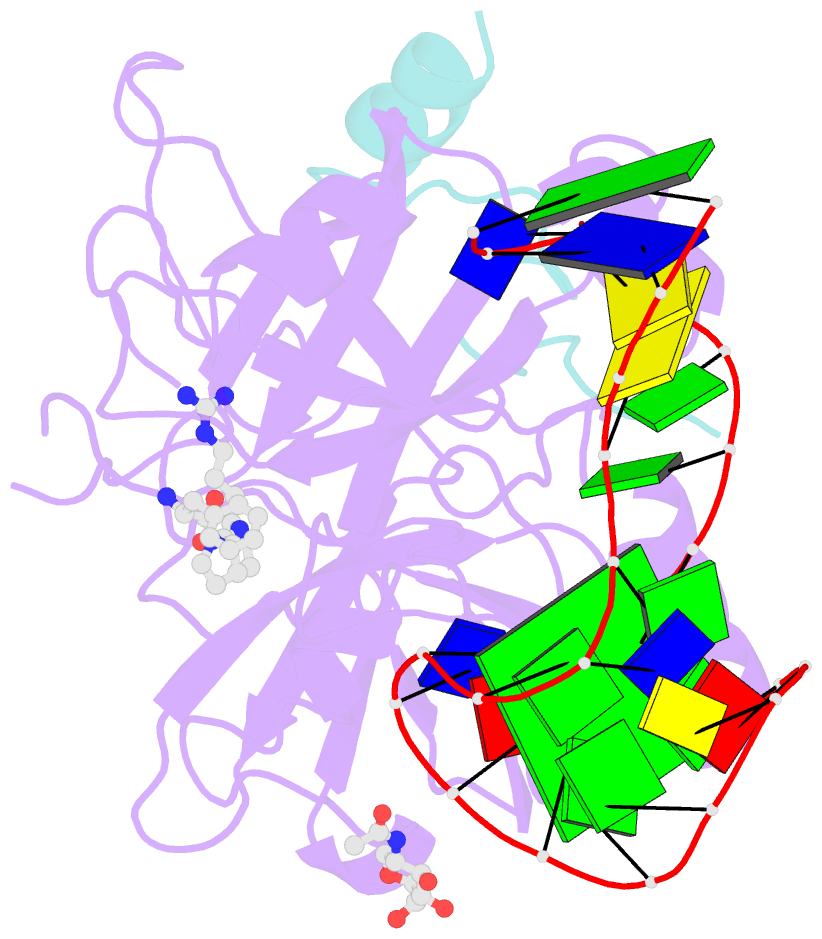
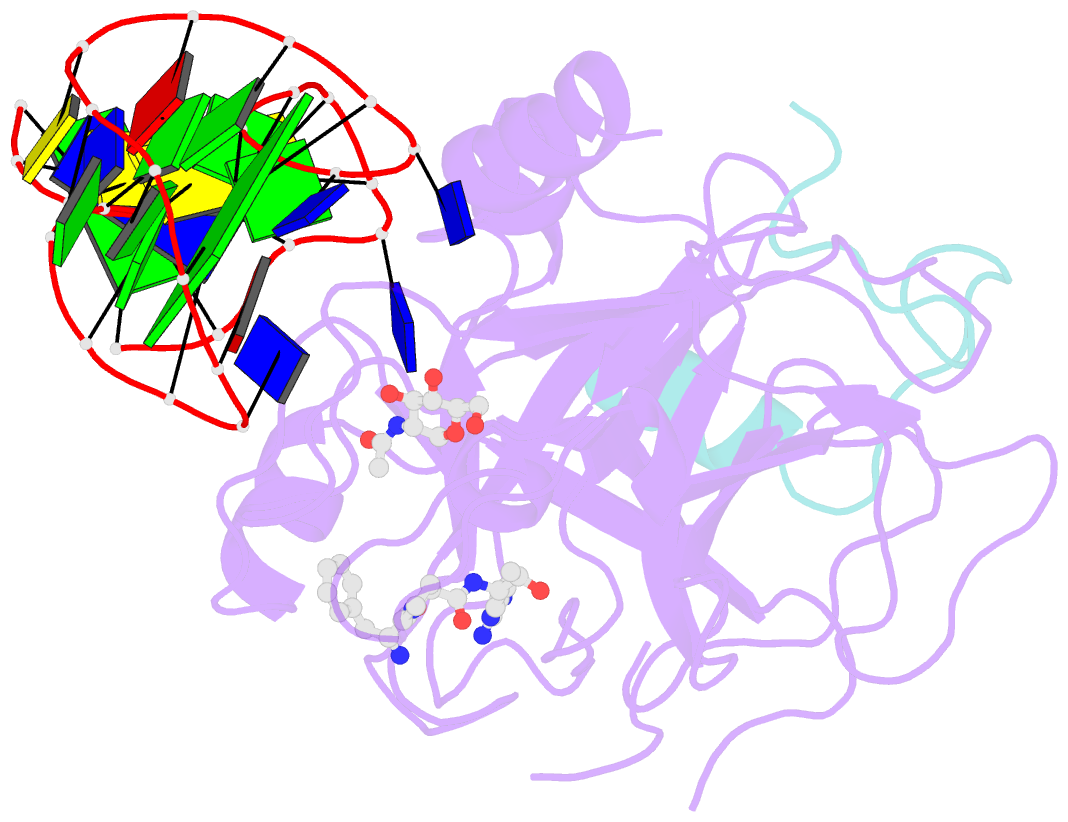
- Crystal structure of human alpha thrombin in complex with a 27-mer aptamer bound to exosite ii
- Reference
- Russo Krauss I, Pica A, Merlino A, Mazzarella L, Sica F (2013): "Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer." Acta Crystallogr.,Sect.D, 69, 2403-2411. doi: 10.1107/S0907444913022269.
- Abstract
- Potent second-generation thrombin aptamers adopt a duplex-quadruplex bimodular folding and recognize thrombin exosite II with very high affinity and specificity. A sound model of these oligonucleotides, either free or in complex with thrombin, is not yet available. Here, a structural study of one of these aptamers, HD22-27mer, is presented. The crystal structure of this aptamer in complex with thrombin displays a novel architecture in which the helical stem is enchained to a pseudo-G-quadruplex. The results also underline the role of the residues that join the duplex and quadruplex motifs and control their recruitment in thrombin binding.