Summary information and primary citation
- PDB-id
- 4iem; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase, lyase-DNA
- Method
- X-ray (2.39 Å)
- Summary
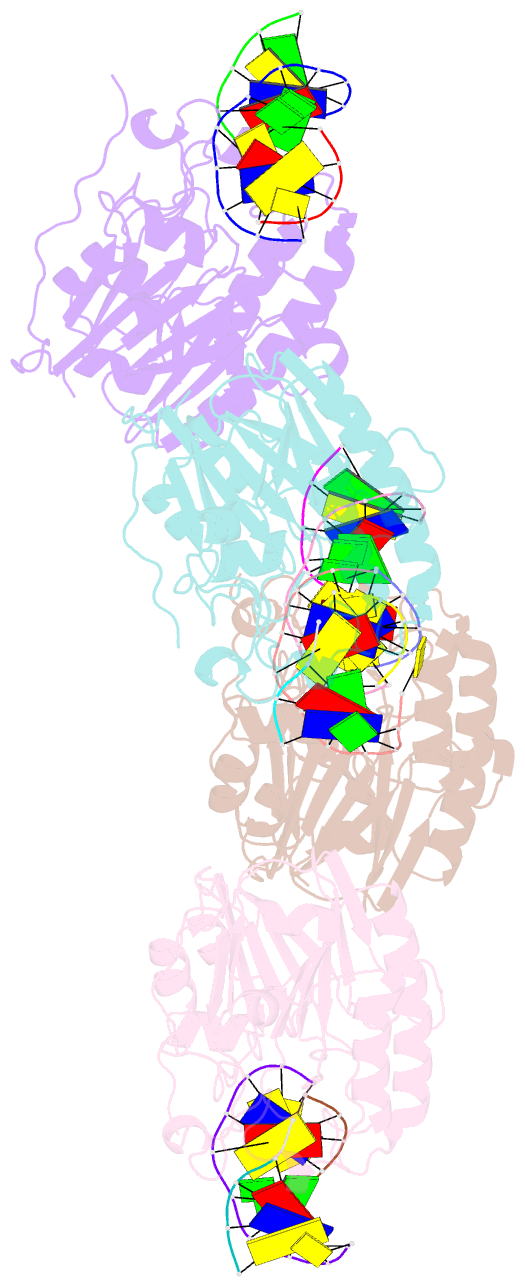
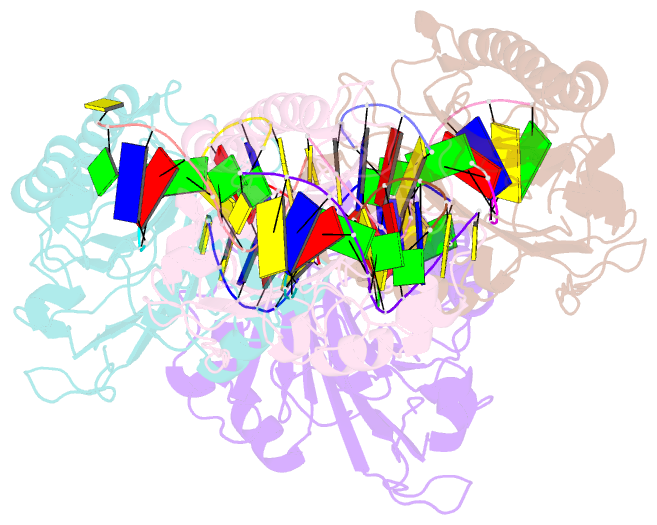
- Human apurinic-apyrimidinic endonuclease (ape1) with product DNA and mg2+
- Reference
- Tsutakawa SE, Shin DS, Mol CD, Izumi T, Arvai AS, Mantha AK, Szczesny B, Ivanov IN, Hosfield DJ, Maiti B, Pique ME, Frankel KA, Hitomi K, Cunningham RP, Mitra S, Tainer JA (2013): "Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes." J.Biol.Chem., 288, 8445-8455. doi: 10.1074/jbc.M112.422774.
- Abstract
- Non-coding apurinic/apyrimidinic (AP) sites in DNA form spontaneously and as DNA base excision repair intermediates are the most common toxic and mutagenic in vivo DNA lesion. For repair, AP sites must be processed by 5' AP endonucleases in initial stages of base repair. Human APE1 and bacterial Nfo represent the two conserved 5' AP endonuclease families in the biosphere; they both recognize AP sites and incise the phosphodiester backbone 5' to the lesion, yet they lack similar structures and metal ion requirements. Here, we determined and analyzed crystal structures of a 2.4 Å resolution APE1-DNA product complex with Mg(2+) and a 0.92 Å Nfo with three metal ions. Structural and biochemical comparisons of these two evolutionarily distinct enzymes characterize key APE1 catalytic residues that are potentially functionally similar to Nfo active site components, as further tested and supported by computational analyses. We observe a magnesium-water cluster in the APE1 active site, with only Glu-96 forming the direct protein coordination to the Mg(2+). Despite differences in structure and metal requirements of APE1 and Nfo, comparison of their active site structures surprisingly reveals strong geometric conservation of the catalytic reaction, with APE1 catalytic side chains positioned analogously to Nfo metal positions, suggesting surprising functional equivalence between Nfo metal ions and APE1 residues. The finding that APE1 residues are positioned to substitute for Nfo metal ions is supported by the impact of mutations on activity. Collectively, the results illuminate the activities of residues, metal ions, and active site features for abasic site endonucleases.