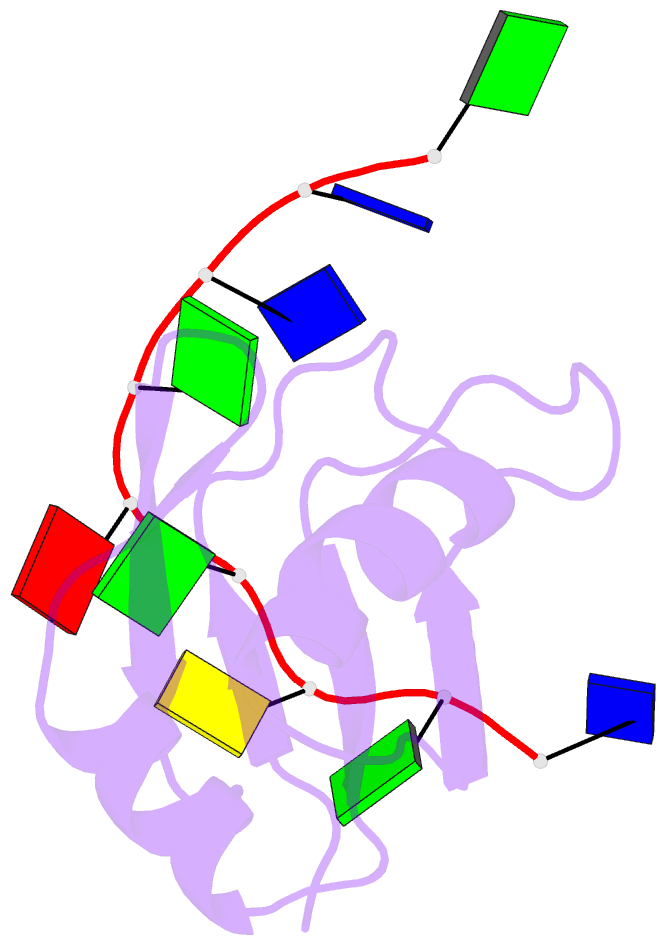
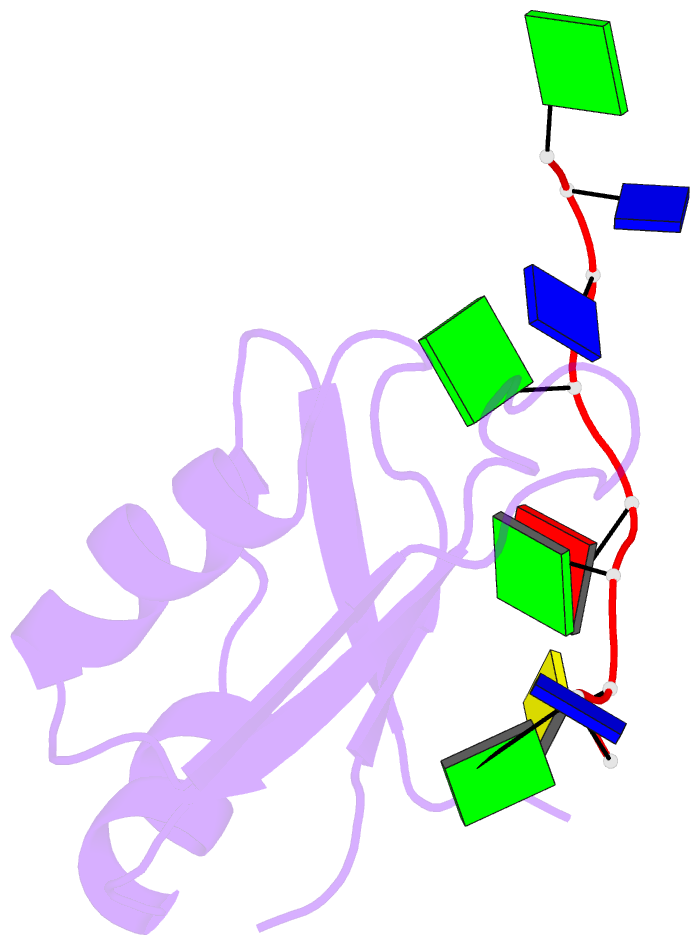
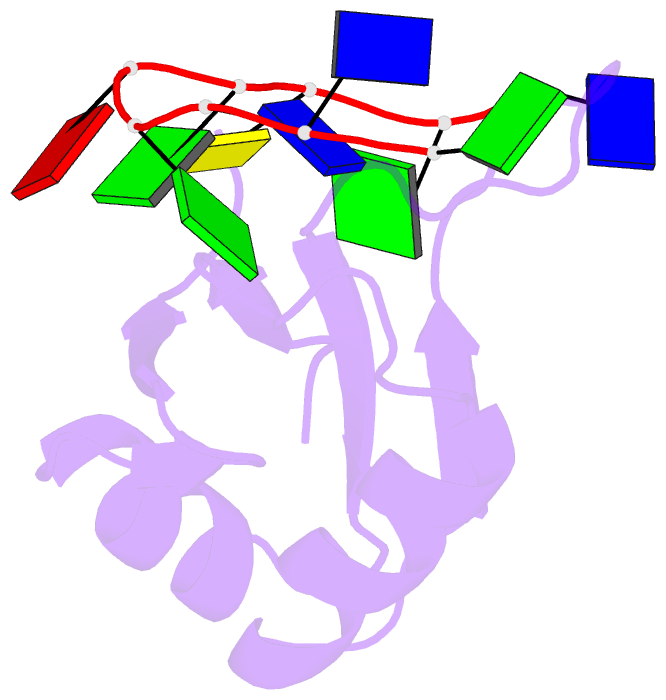
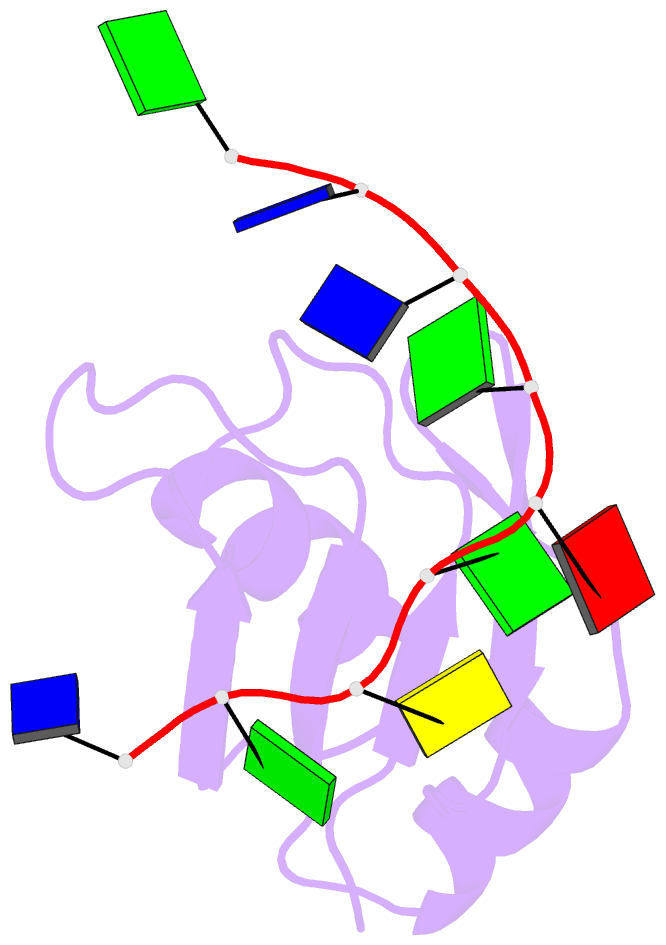
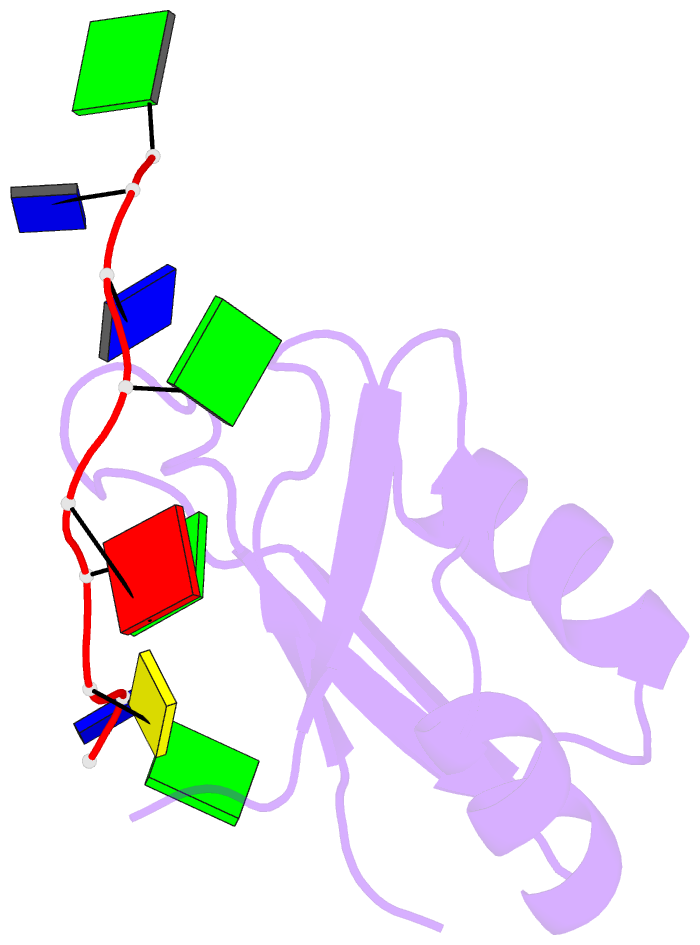
Summary information and primary citation
- PDB-id
- 4iuf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription regulator-DNA
- Method
- X-ray (2.752 Å)
- Summary
- Crystal structure of human tdp-43 rrm1 domain in complex with a single-stranded DNA
- Reference
- Kuo PH, Chiang CH, Wang YT, Doudeva LG, Yuan HS (2014): "The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids." Nucleic Acids Res., 42, 4712-4722. doi: 10.1093/nar/gkt1407.
- Abstract
- TDP-43 is an important pathological protein that aggregates in the diseased neuronal cells and is linked to various neurodegenerative disorders. In normal cells, TDP-43 is primarily an RNA-binding protein; however, how the dimeric TDP-43 binds RNA via its two RNA recognition motifs, RRM1 and RRM2, is not clear. Here we report the crystal structure of human TDP-43 RRM1 in complex with a single-stranded DNA showing that RRM1 binds the nucleic acid extensively not only by the conserved β-sheet residues but also by the loop residues. Mutational and biochemical assays further reveal that both RRMs in TDP-43 dimers participate in binding of UG-rich RNA or TG-rich DNA with RRM1 playing a dominant role and RRM2 playing a supporting role. Moreover, RRM1 of the amyotrophic lateral sclerosis-linked mutant D169G binds DNA as efficiently as the wild type; nevertheless, it is more resistant to thermal denaturation, suggesting that the resistance to degradation is likely linked to TDP-43 proteinopathies. Taken together all the data, we suggest a model showing that the two RRMs in each protomer of TDP-43 homodimer work together in RNA binding and thus the dimeric TDP-43 recognizes long clusters of UG-rich RNA to achieve high affinity and specificity.