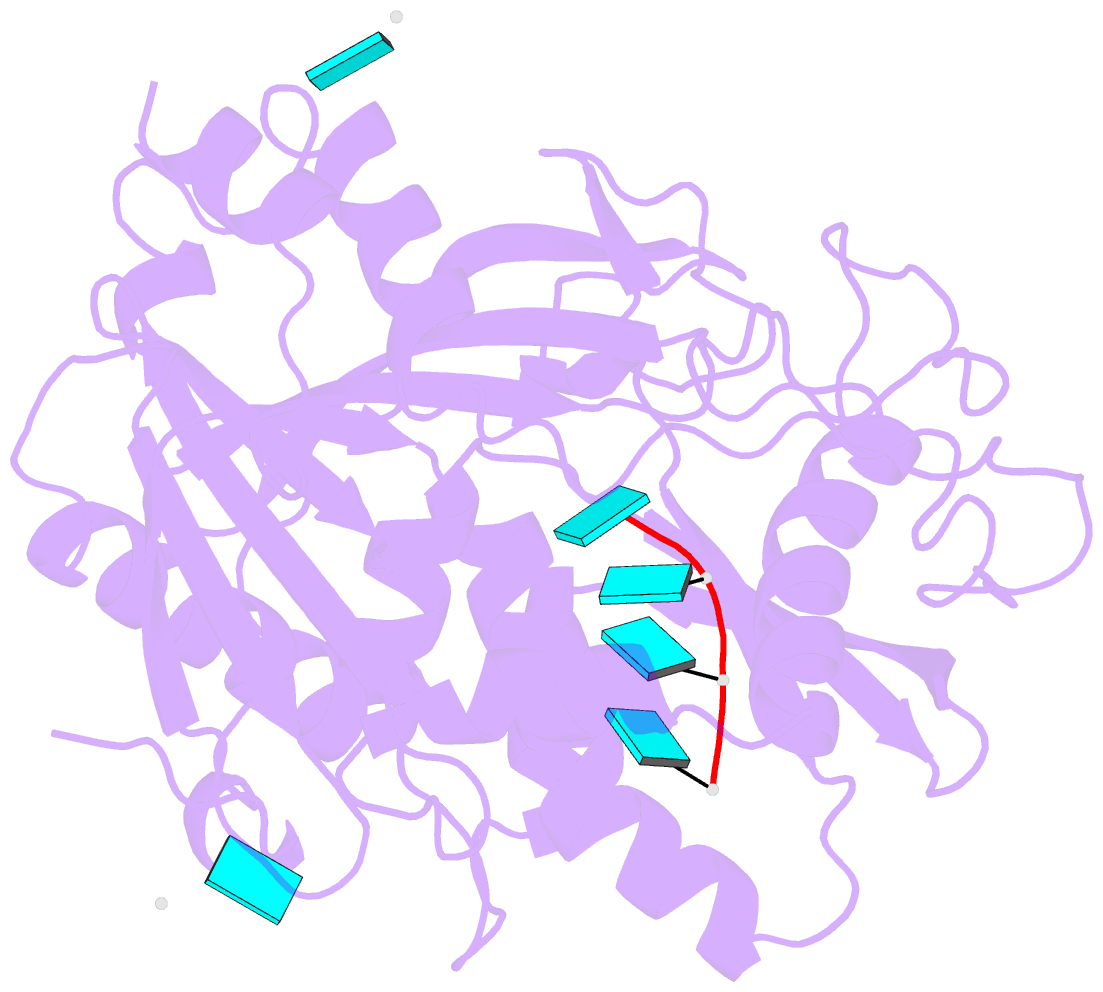
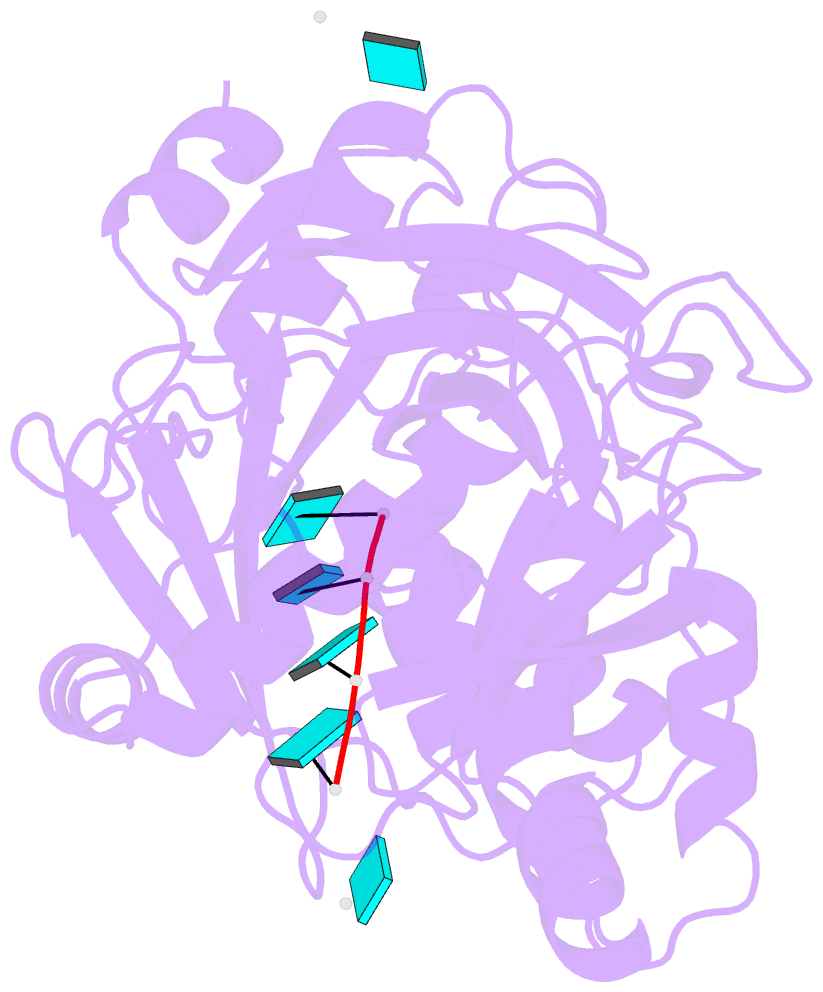
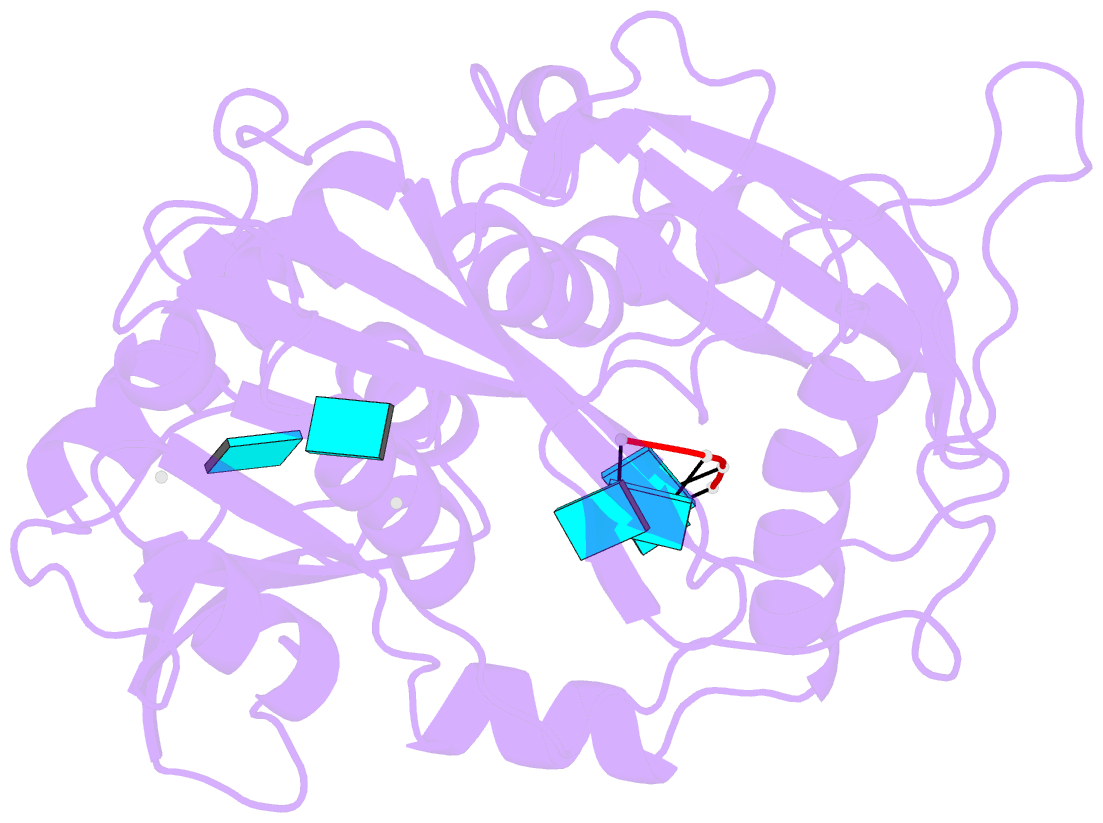
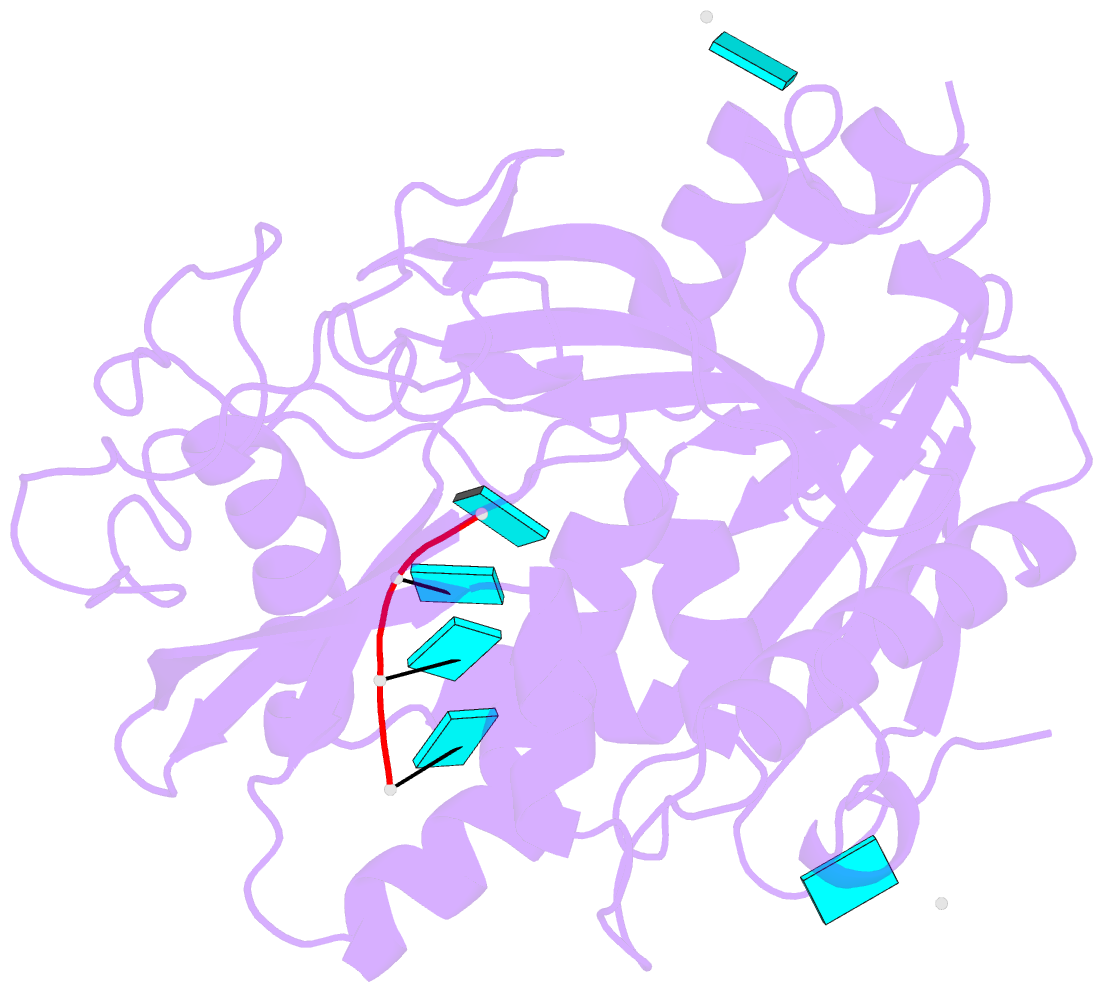
Summary information and primary citation
- PDB-id
-
4j7m;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (1.7 Å)
- Summary
- Crystal structure of mouse dxo in complex with
substrate mimic RNA and calcium ion
- Reference
-
Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M (2013):
"A
mammalian pre-mRNA 5' end capping quality control
mechanism and an unexpected link of capping to pre-mRNA
processing." Mol.Cell, 50,
104-115. doi: 10.1016/j.molcel.2013.02.017.
- Abstract
- Recently, we reported that two homologous yeast
proteins, Rai1 and Dxo1, function in a quality control
mechanism to clear cells of incompletely 5' end-capped
messenger RNAs (mRNAs). Here, we report that their
mammalian homolog, Dom3Z (referred to as DXO), possesses
pyrophosphohydrolase, decapping, and 5'-to-3'
exoribonuclease activities. Surprisingly, we found that DXO
preferentially degrades defectively capped pre-mRNAs in
cells. Additional studies show that incompletely capped
pre-mRNAs are inefficiently spliced at all introns, a fact
that contrasts with current understanding, and are also
poorly cleaved for polyadenylation. Crystal structures of
DXO in complex with substrate mimic and products at a
resolution of up to 1.5Å provide elegant insights into the
catalytic mechanism and molecular basis for their three
apparently distinct activities. Our data reveal a pre-mRNA
5' end capping quality control mechanism in mammalian
cells, indicating DXO as the central player for this
mechanism, and demonstrate an unexpected intimate link
between proper 5' end capping and subsequent pre-mRNA
processing.