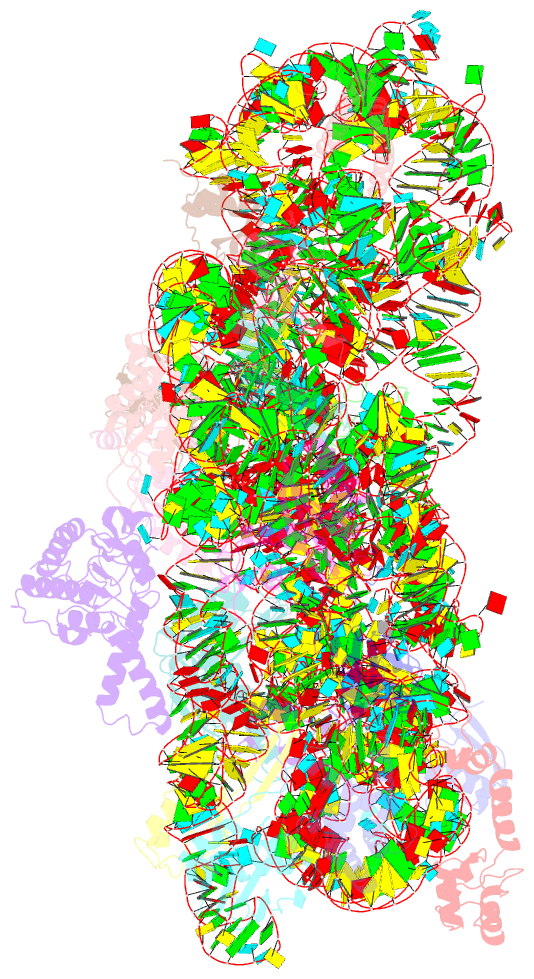
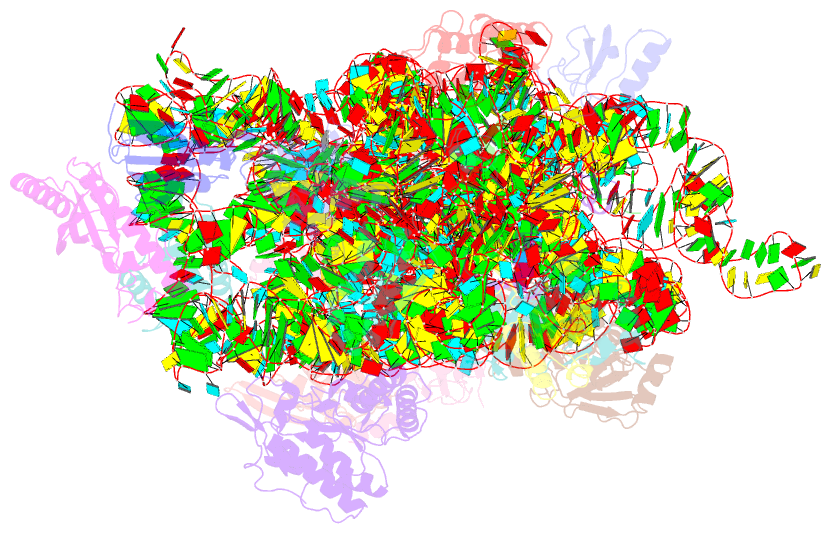
Summary information and primary citation
- PDB-id
- 4ji4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome-antibiotic
- Method
- X-ray (3.692 Å)
- Summary
- Crystal structure of 30s ribosomal subunit from thermus thermophilus
- Reference
- Demirci H, Wang L, Murphy FV, Murphy EL, Carr JF, Blanchard SC, Jogl G, Dahlberg AE, Gregory ST (2013): "The central role of protein S12 in organizing the structure of the decoding site of the ribosome." Rna, 19, 1791-1801. doi: 10.1261/rna.040030.113.
- Abstract
- The ribosome decodes mRNA by monitoring the geometry of codon-anticodon base-pairing using a set of universally conserved 16S rRNA nucleotides within the conformationally dynamic decoding site. By applying single-molecule FRET and X-ray crystallography, we have determined that conditional-lethal, streptomycin-dependence mutations in ribosomal protein S12 interfere with tRNA selection by allowing conformational distortions of the decoding site that impair GTPase activation of EF-Tu during the tRNA selection process. Distortions in the decoding site are reversed by streptomycin or by a second-site suppressor mutation in 16S rRNA. These observations encourage a refinement of the current model for decoding, wherein ribosomal protein S12 and the decoding site collaborate to optimize codon recognition and substrate discrimination during the early stages of the tRNA selection process.