Summary information and primary citation
- PDB-id
- 4jng; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (2.12 Å)
- Summary
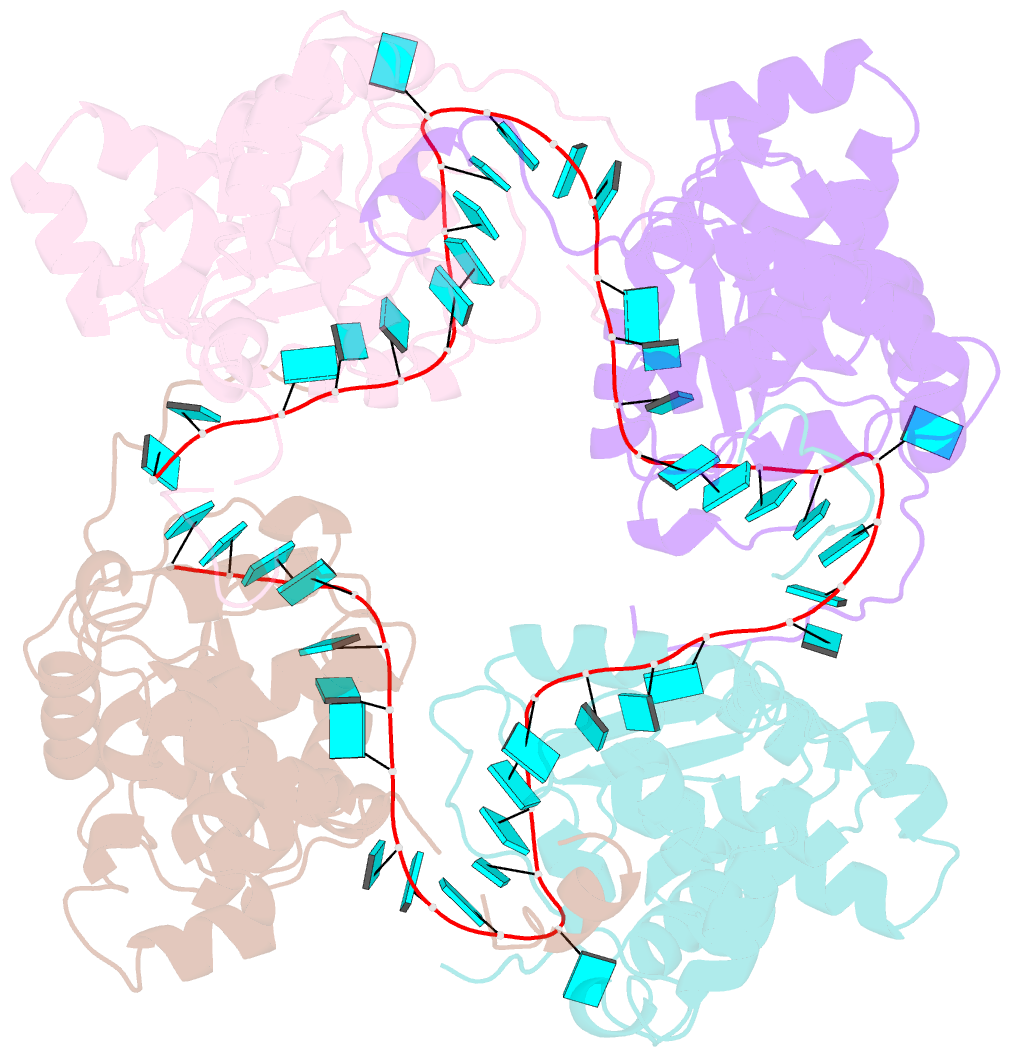
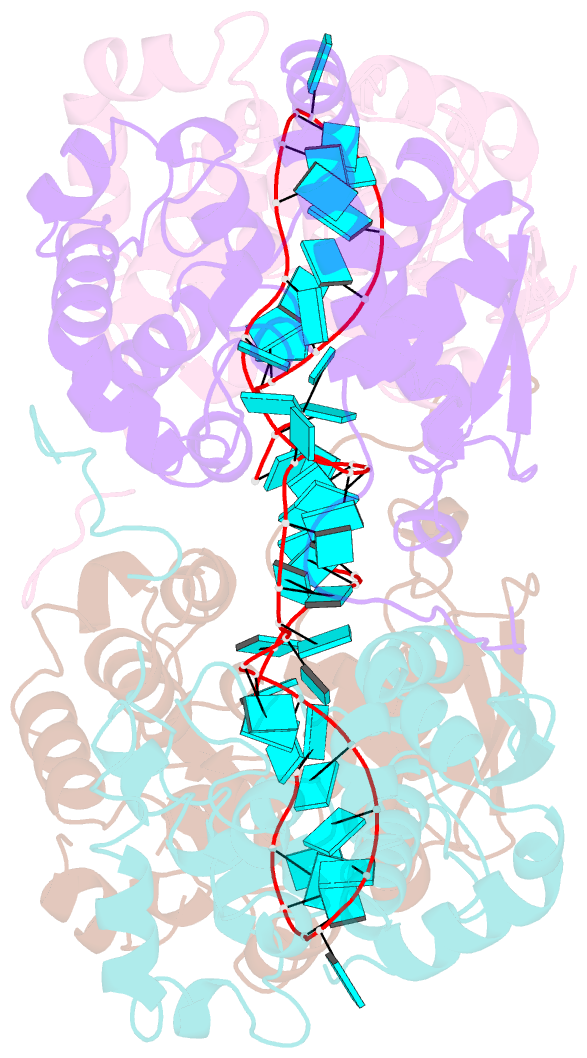
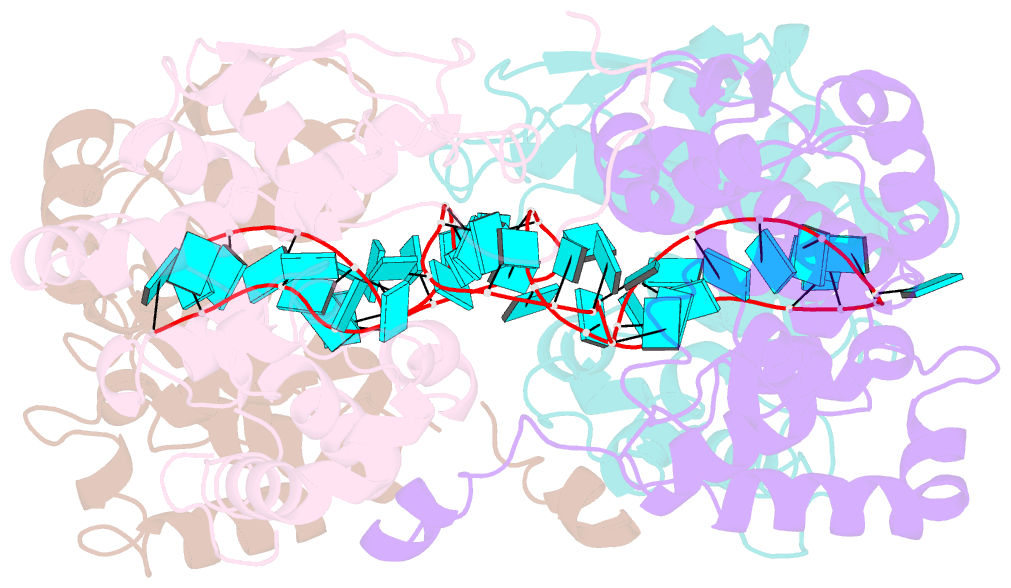
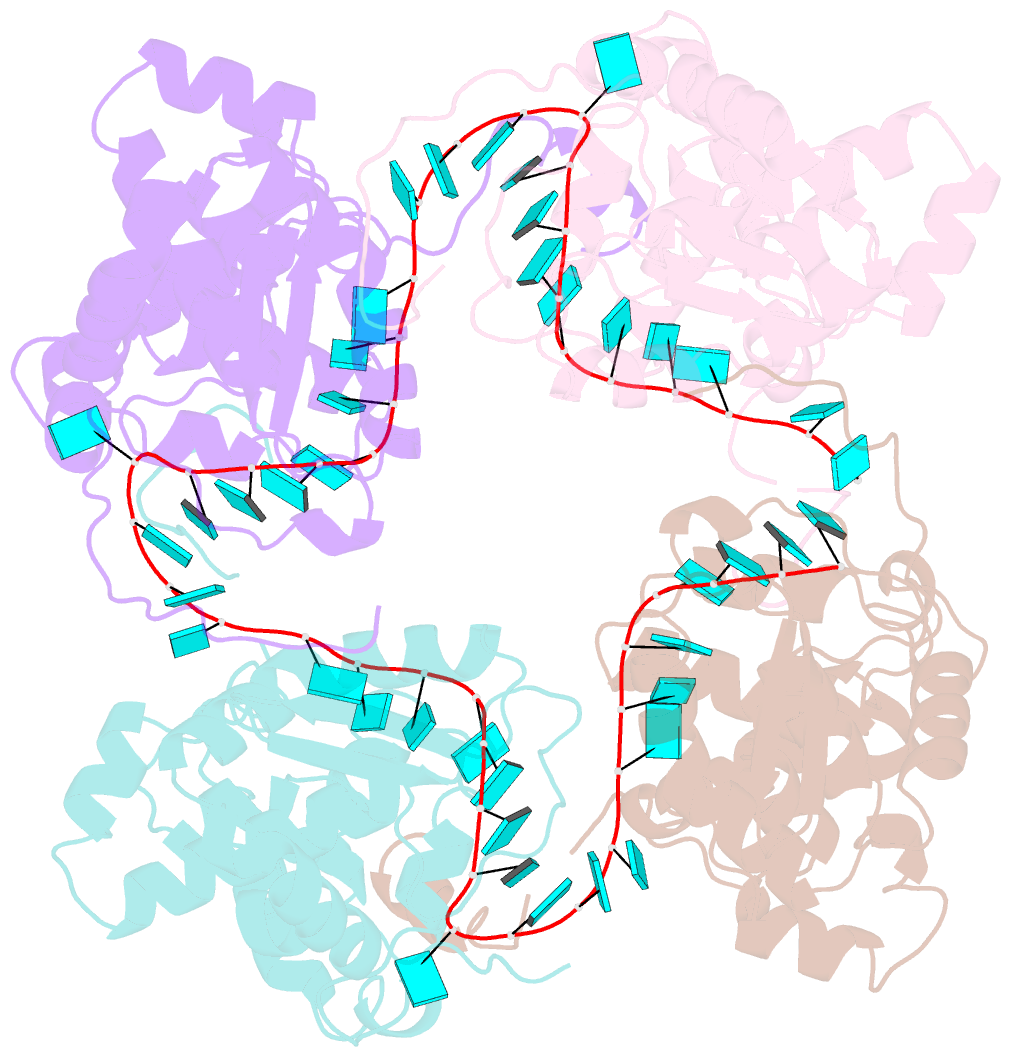
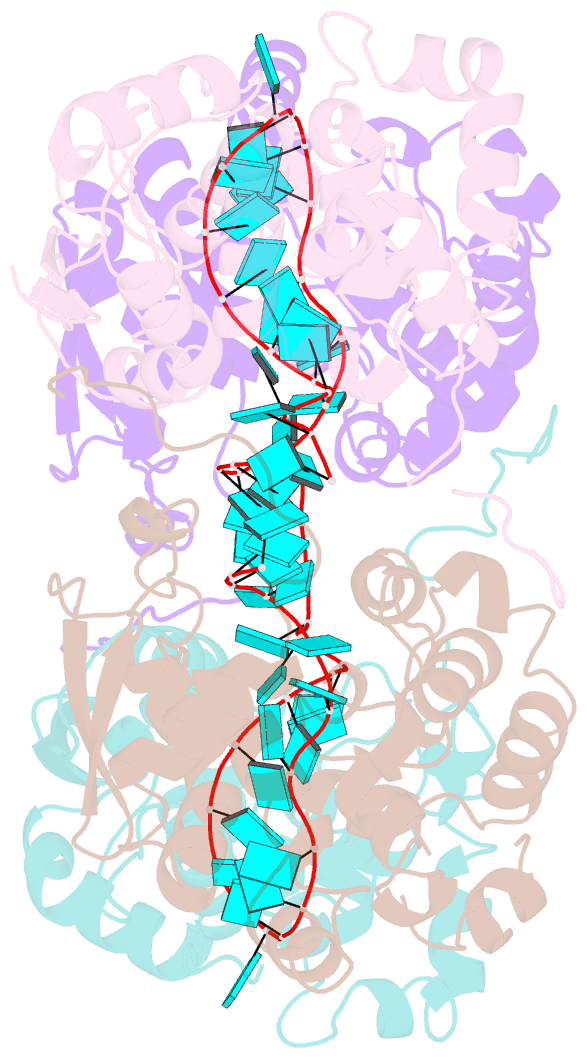
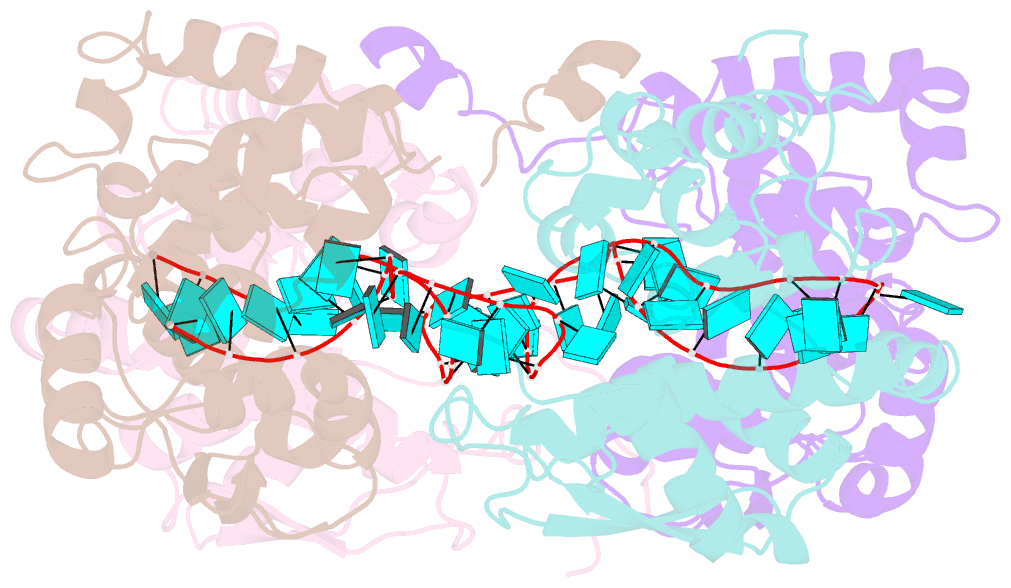
- Schmallenberg virus nucleoprotein-RNA complex
- Reference
- Dong HH, Li P, Bottcher B, Elliott RM, Dong CJ (2013): "Crystal structure of Schmallenberg orthobunyavirus nucleoprotein-RNA complex reveals a novel RNA sequestration mechanism." Rna, 19, 1129-1136. doi: 10.1261/rna.039057.113.
- Abstract
- Schmallenberg virus (SBV) is a newly emerged orthobunyavirus (family Bunyaviridae) that has caused severe disease in the offspring of farm animals across Europe. Like all orthobunyaviruses, SBV contains a tripartite negative-sense RNA genome that is encapsidated by the viral nucleocapsid (N) protein in the form of a ribonucleoprotein complex (RNP). We recently reported the three-dimensional structure of SBV N that revealed a novel fold. Here we report the crystal structure of the SBV N protein in complex with a 42-nt-long RNA to 2.16 Å resolution. The complex comprises a tetramer of N that encapsidates the RNA as a cross-shape inside the protein ring structure, with each protomer bound to 11 ribonucleotides. Eight bases are bound in the positively charged cleft between the N- and C-terminal domains of N, and three bases are shielded by the extended N-terminal arm. SBV N appears to sequester RNA using a different mechanism compared with the nucleoproteins of other negative-sense RNA viruses. Furthermore, the structure suggests that RNA binding results in conformational changes of some residues in the RNA-binding cleft and the N- and C-terminal arms. Our results provide new insights into the novel mechanism of RNA encapsidation by orthobunyaviruses.