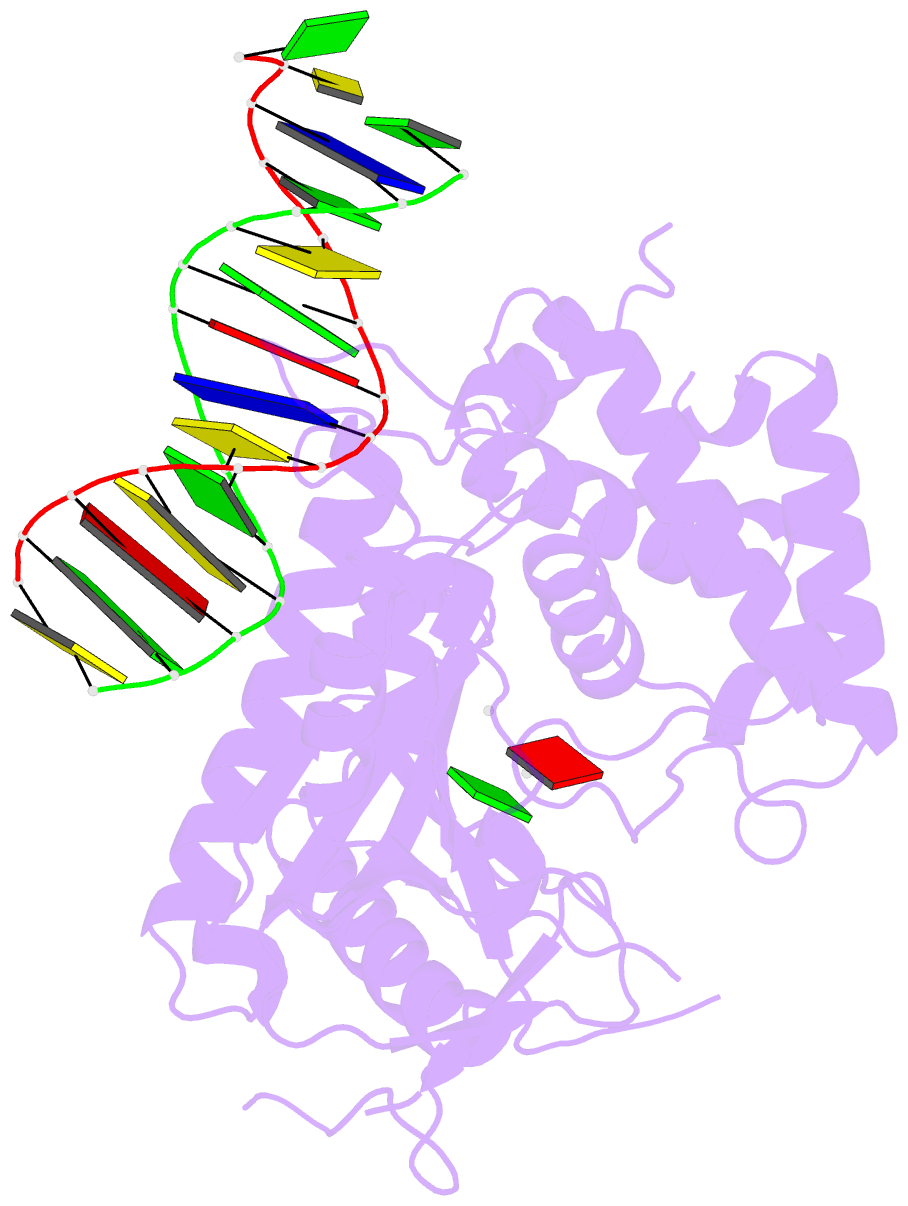
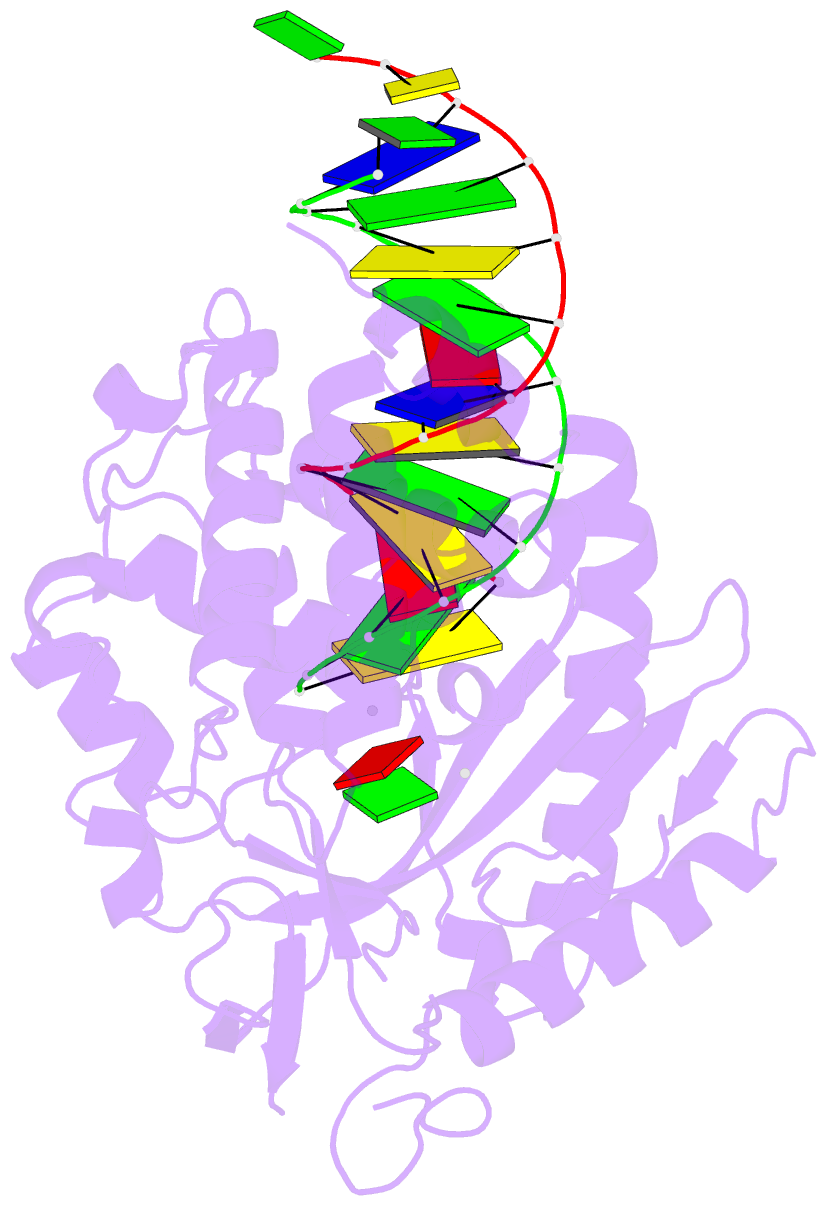
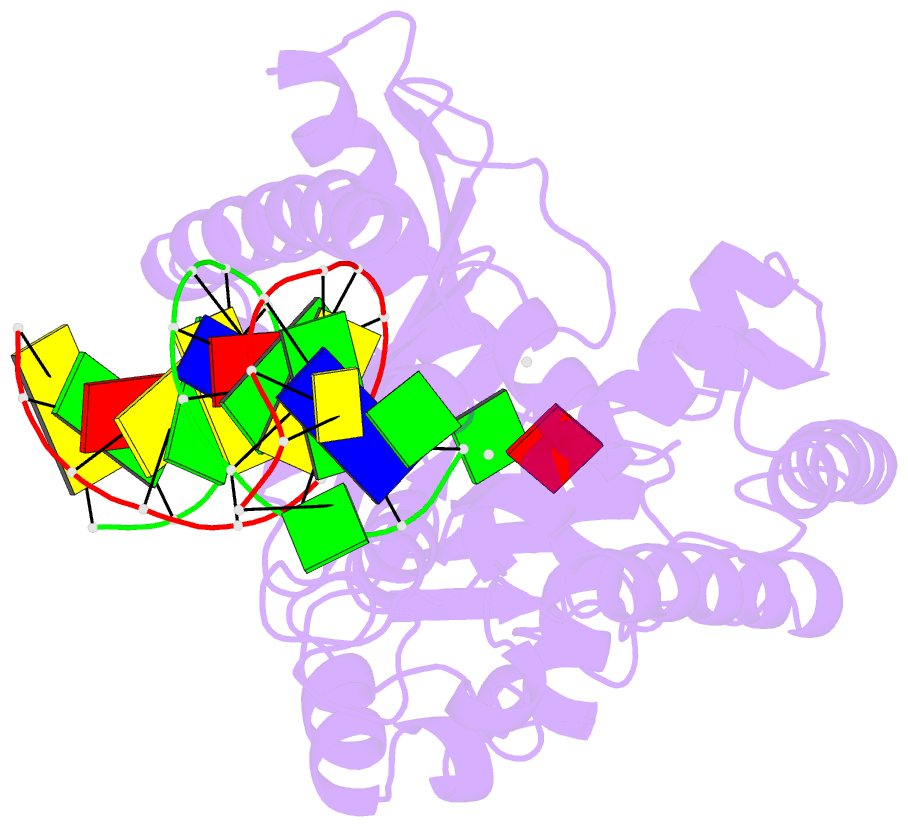
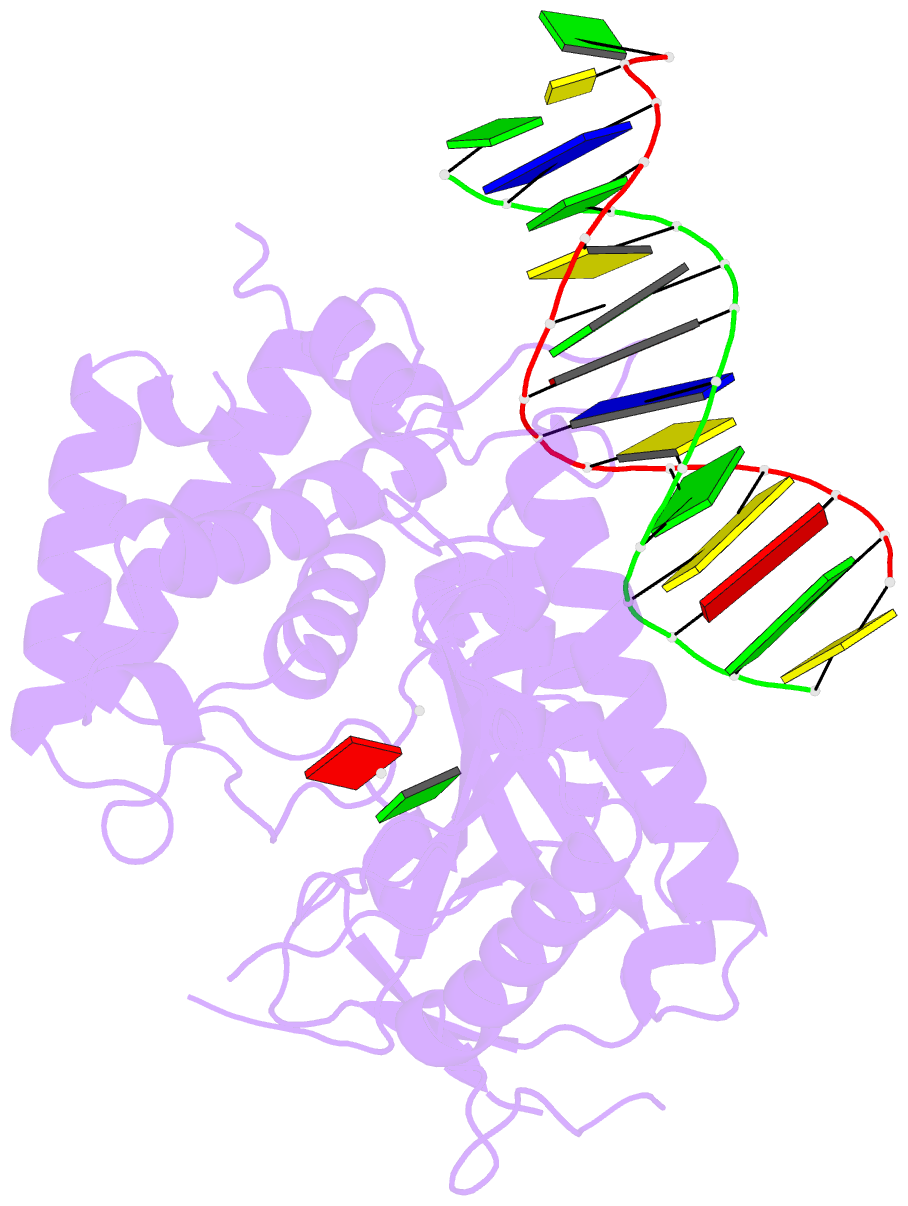
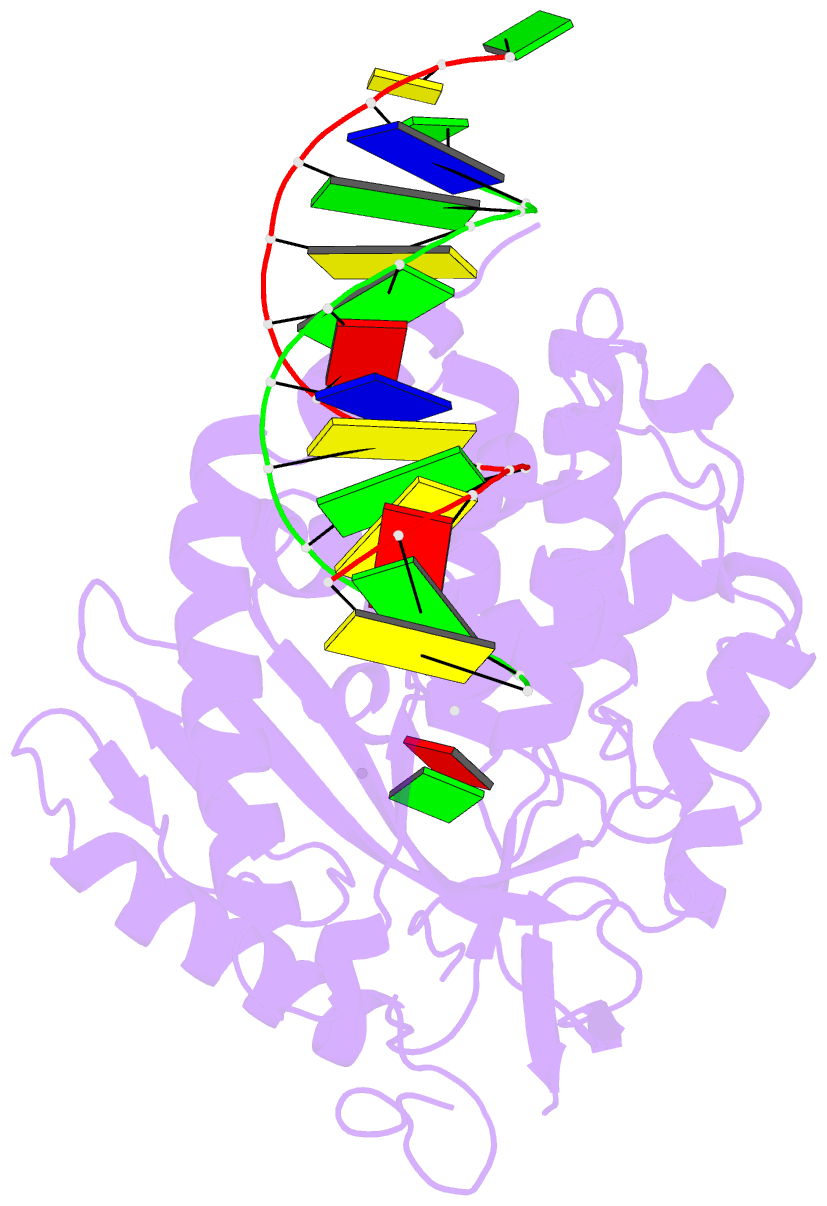
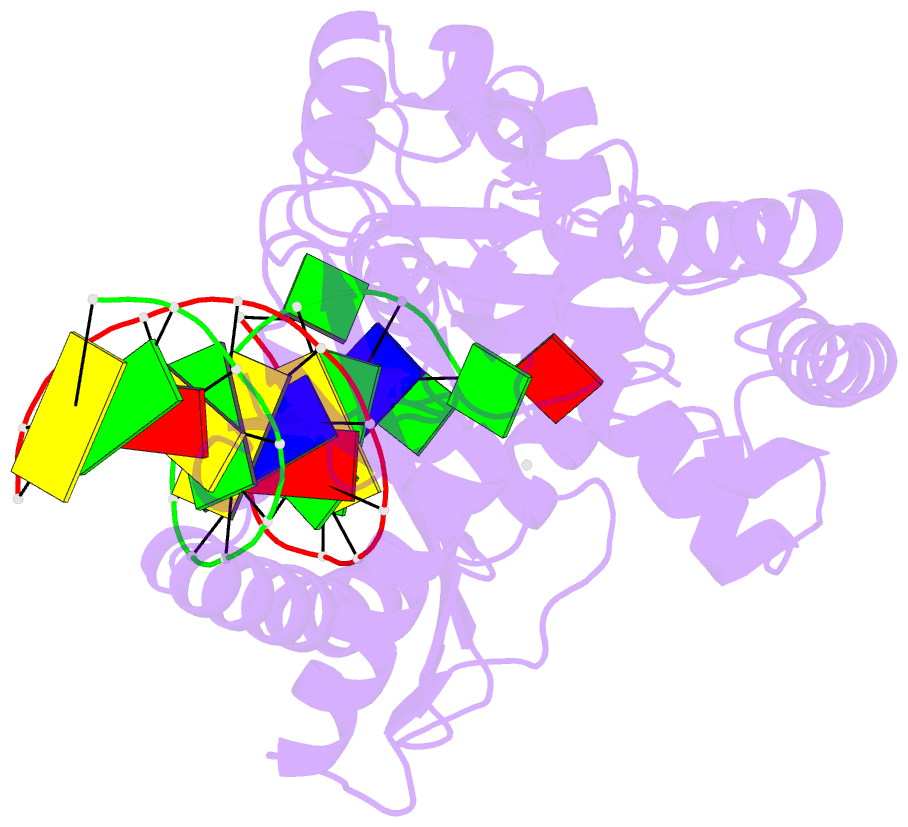
Summary information and primary citation
- PDB-id
- 4kb6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (3.08 Å)
- Summary
- Structure of porcine cyclic gmp amp synthase (cgas) in complex with DNA, atp and gtp
- Reference
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP (2013): "Structural mechanism of cytosolic DNA sensing by cGAS." Nature, 498, 332-337. doi: 10.1038/nature12305.
- Abstract
- Cytosolic DNA arising from intracellular bacterial or viral infections is a powerful pathogen-associated molecular pattern (PAMP) that leads to innate immune host defence by the production of type I interferon and inflammatory cytokines. Recognition of cytosolic DNA by the recently discovered cyclic-GMP-AMP (cGAMP) synthase (cGAS) induces the production of cGAMP to activate the stimulator of interferon genes (STING). Here we report the crystal structure of cGAS alone and in complex with DNA, ATP and GTP along with functional studies. Our results explain the broad DNA sensing specificity of cGAS, show how cGAS catalyses dinucleotide formation and indicate activation by a DNA-induced structural switch. cGAS possesses a remarkable structural similarity to the antiviral cytosolic double-stranded RNA sensor 2'-5'oligoadenylate synthase (OAS1), but contains a unique zinc thumb that recognizes B-form double-stranded DNA. Our results mechanistically unify dsRNA and dsDNA innate immune sensing by OAS1 and cGAS nucleotidyl transferases.