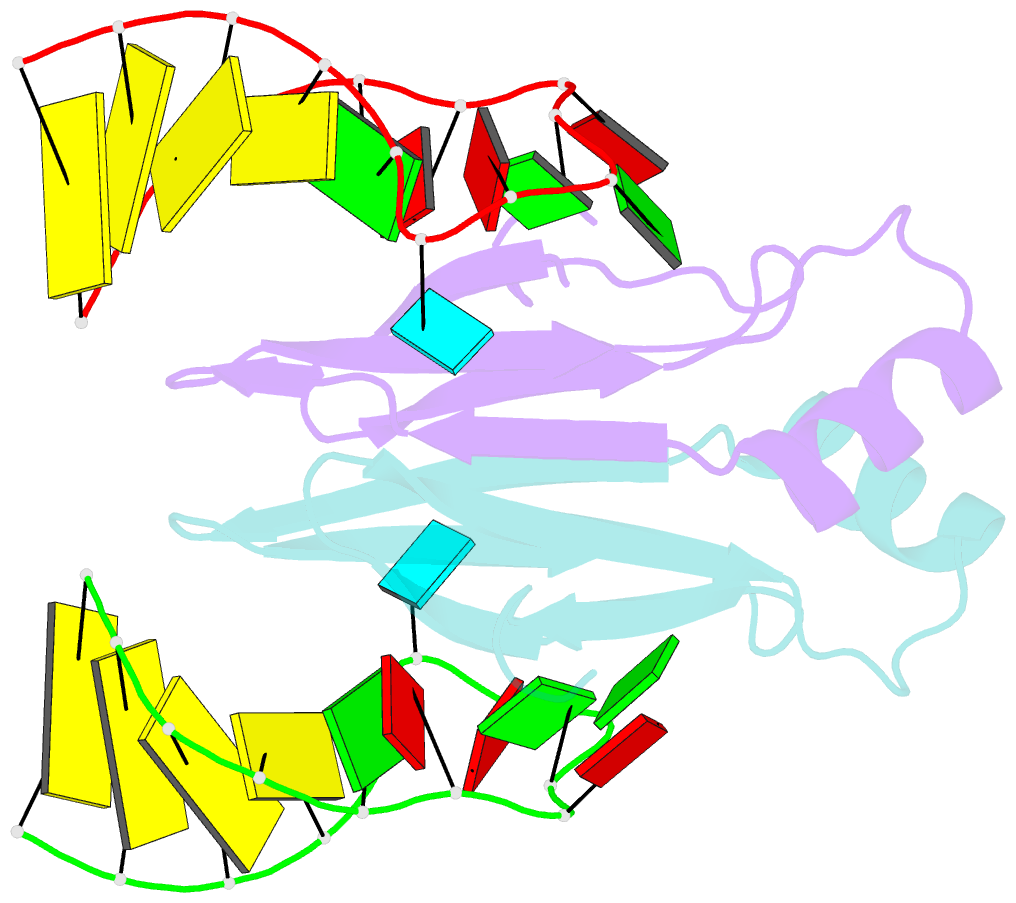
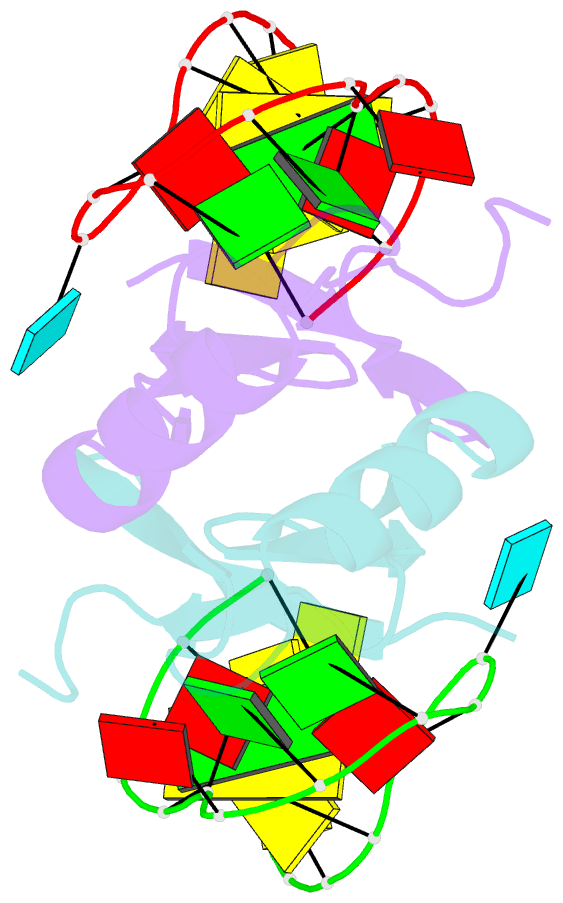
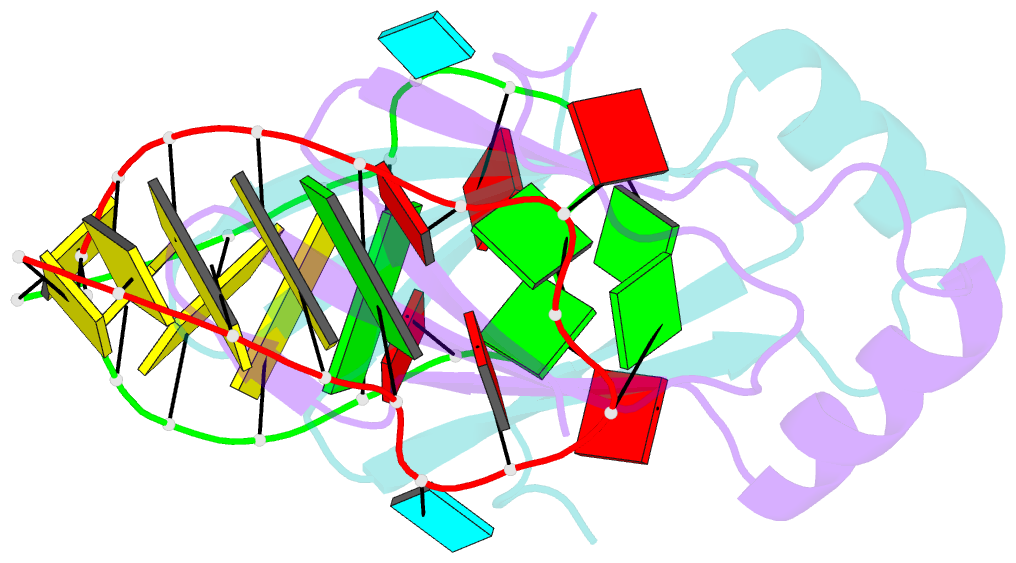
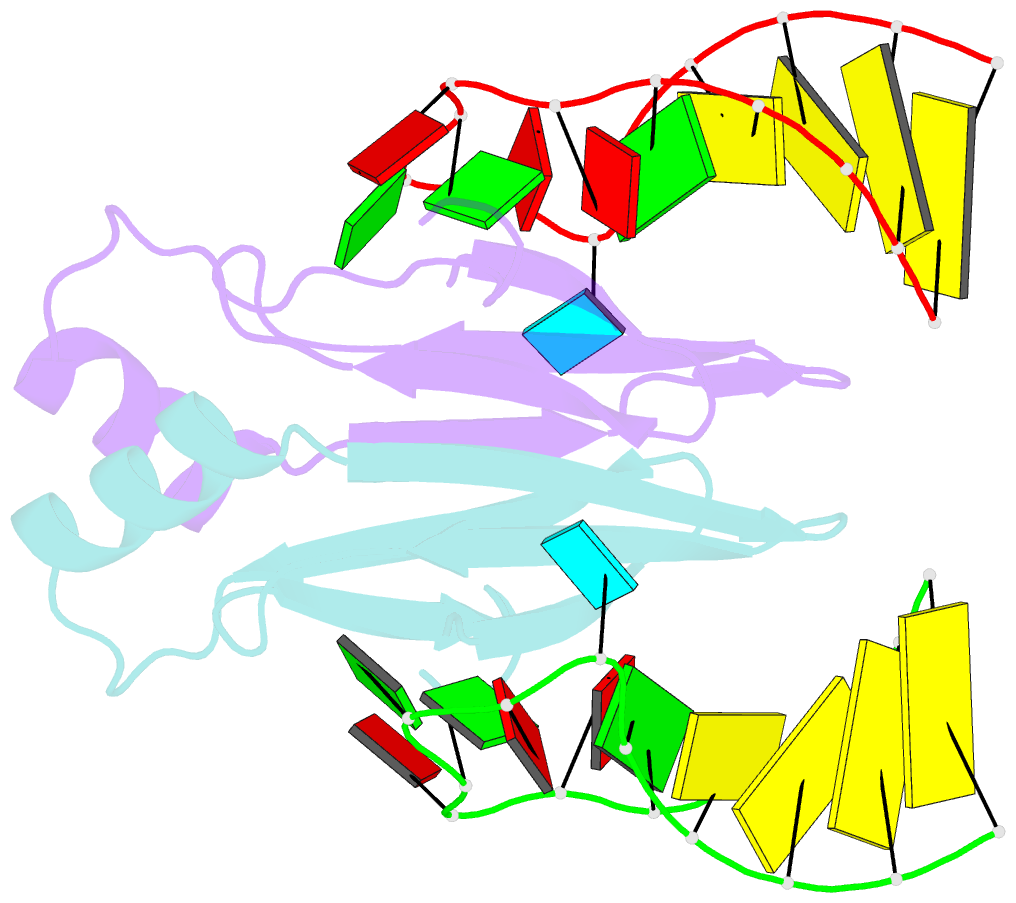
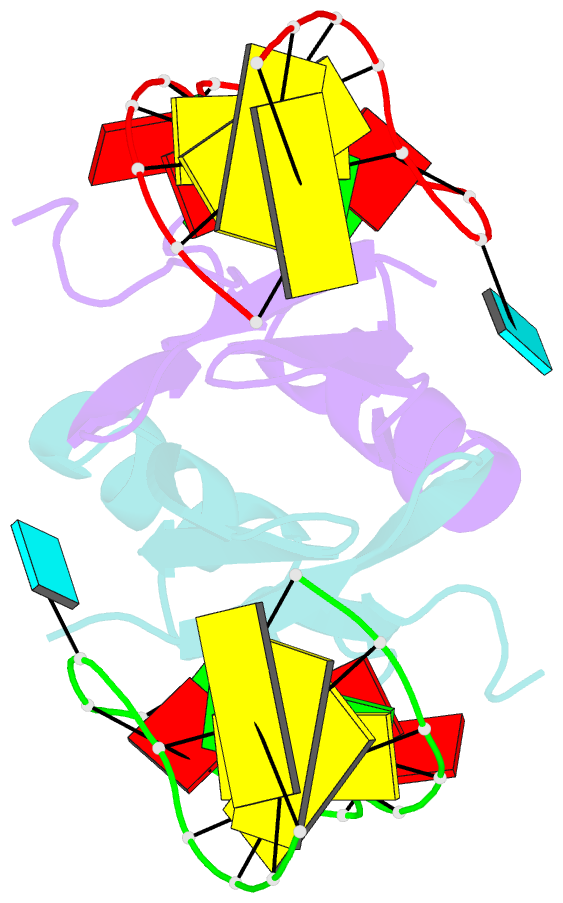
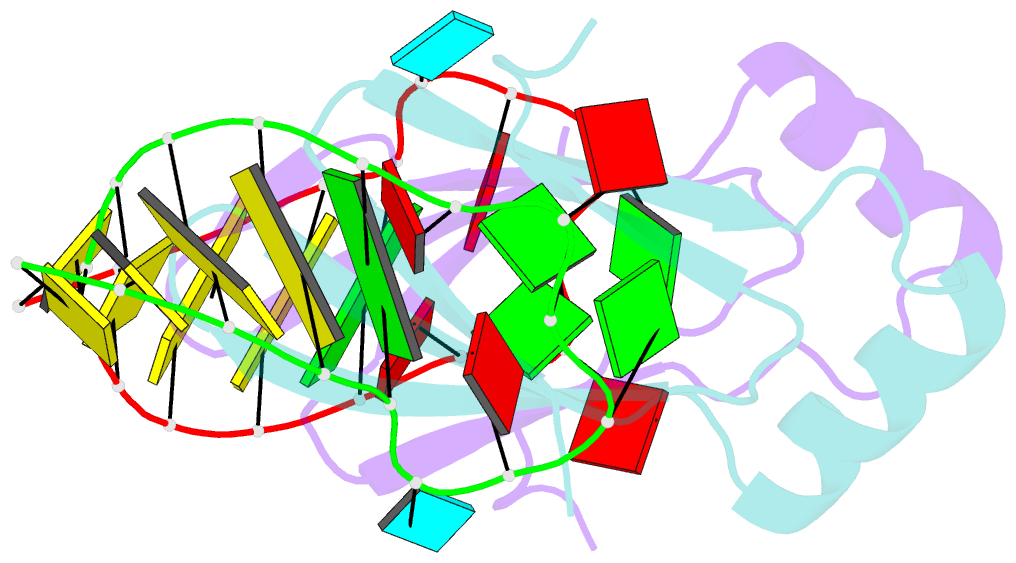
Summary information and primary citation
- PDB-id
- 4kji; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.2 Å)
- Summary
- Novel re-arrangement of an rsma-csra family protein to create a structurally distinct new RNA-binding family member
- Reference
- Morris ER, Hall G, Li C, Heeb S, Kulkarni RV, Lovelock L, Silistre H, Messina M, Camara M, Emsley J, Williams P, Searle MS (2013): "Structural Rearrangement in an RsmA/CsrA Ortholog of Pseudomonas aeruginosa Creates a Dimeric RNA-Binding Protein, RsmN." Structure, 21, 1659-1671. doi: 10.1016/j.str.2013.07.007.
- Abstract
- In bacteria, the highly conserved RsmA/CsrA family of RNA-binding proteins functions as global posttranscriptional regulators acting on mRNA translation and stability. Through phenotypic complementation of an rsmA mutant in Pseudomonas aeruginosa, we discovered a family member, termed RsmN. Elucidation of the RsmN crystal structure and that of the complex with a hairpin from the sRNA, RsmZ, reveals a uniquely inserted α helix, which redirects the polypeptide chain to form a distinctly different protein fold to the domain-swapped dimeric structure of RsmA homologs. The overall β sheet structure required for RNA recognition is, however, preserved with compensatory sequence and structure differences, allowing the RsmN dimer to target binding motifs in both structured hairpin loops and flexible disordered RNAs. Phylogenetic analysis indicates that, although RsmN appears unique to P. aeruginosa, homologous proteins with the inserted α helix are more widespread and arose as a consequence of a gene duplication event.