Summary information and primary citation
- PDB-id
- 4l8r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-RNA binding protein-hydrolase
- Method
- X-ray (2.6 Å)
- Summary
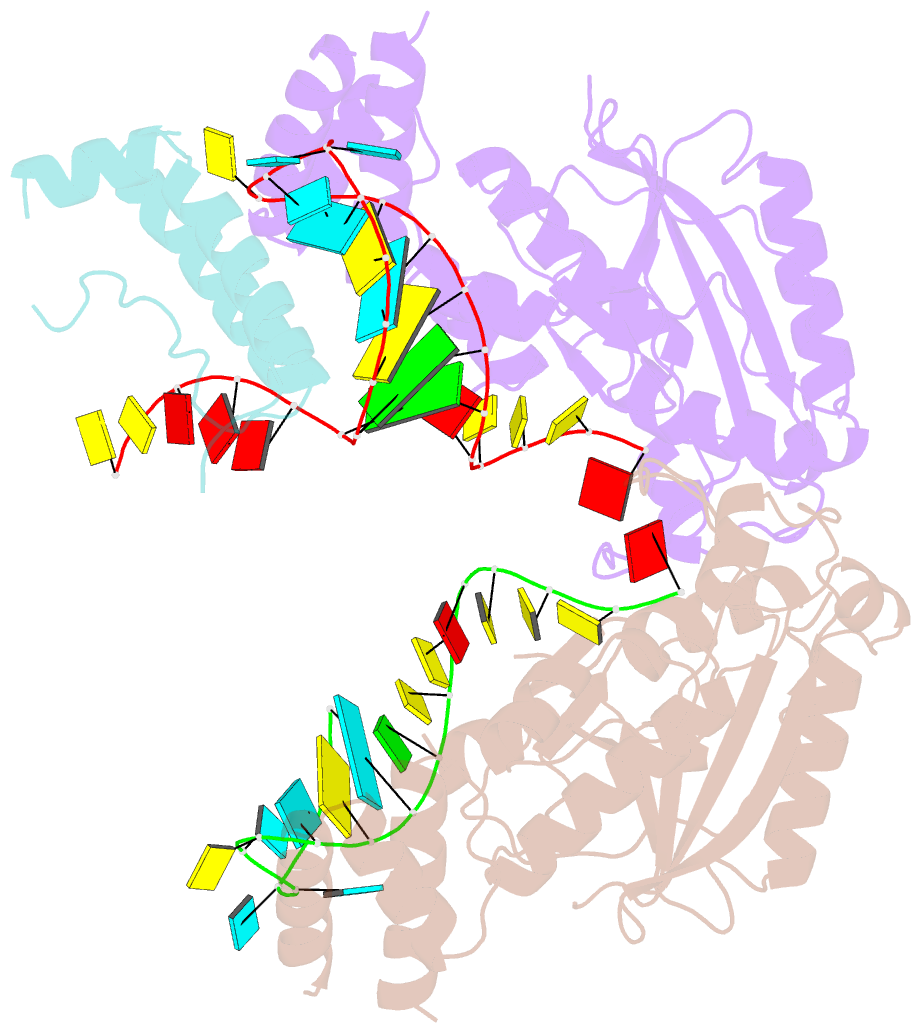
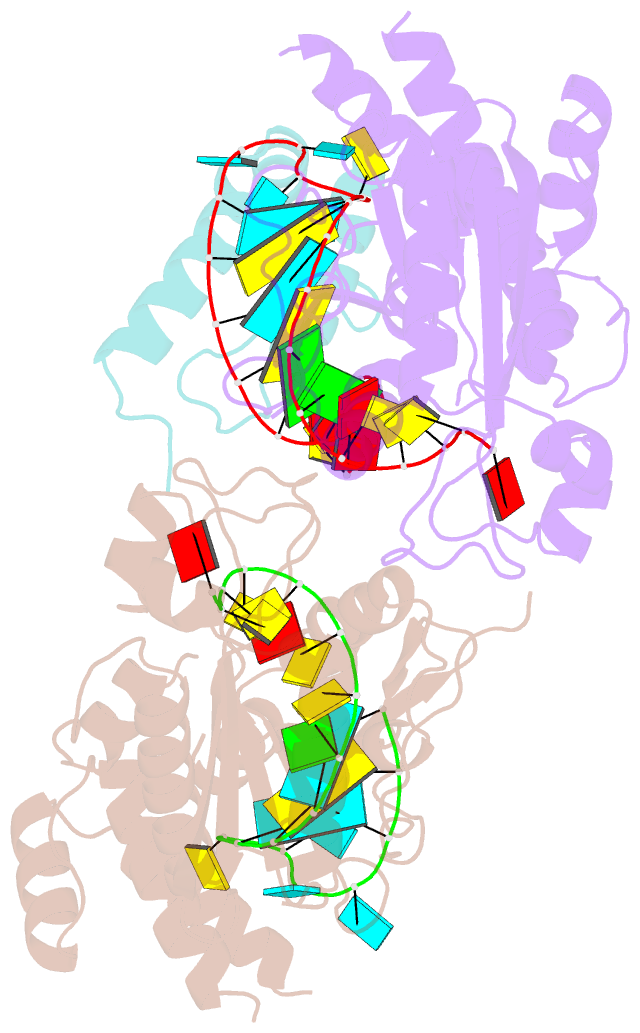
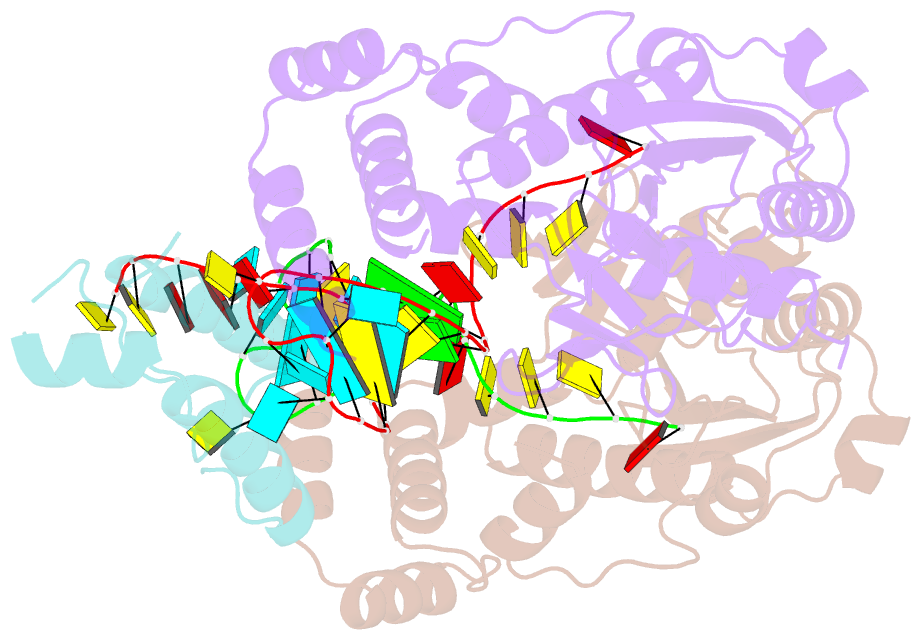
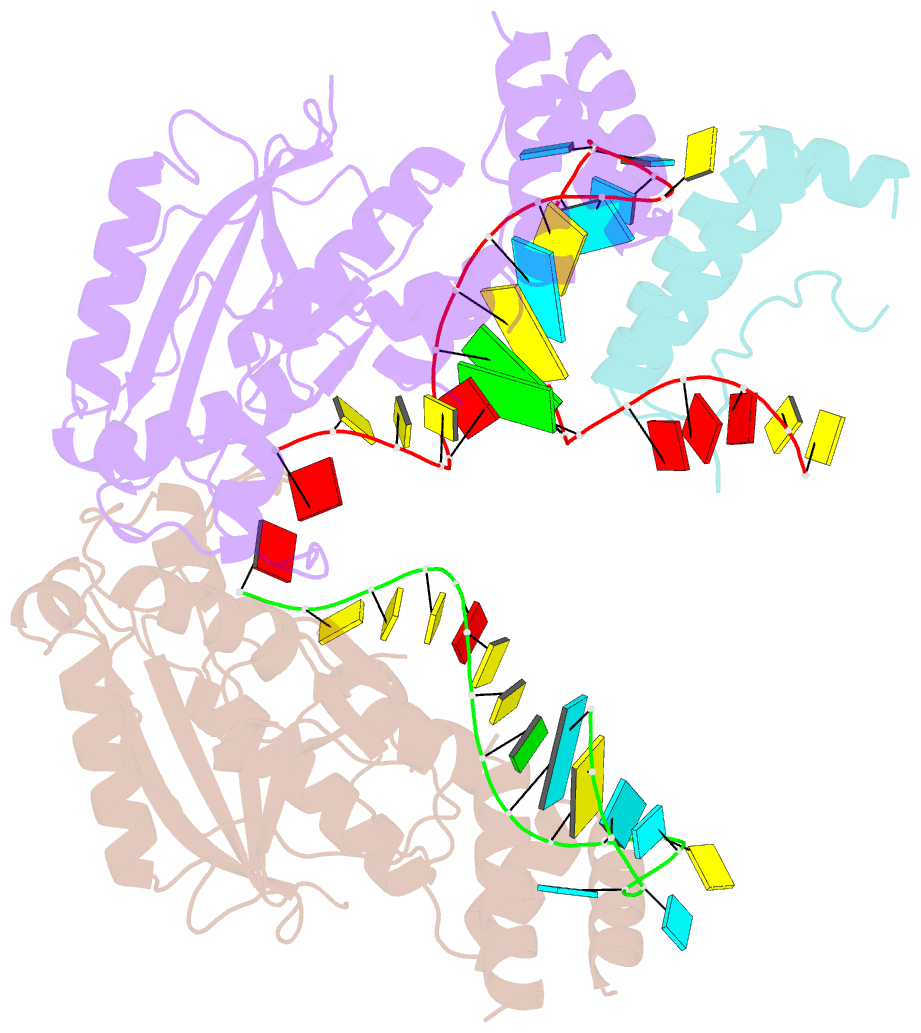
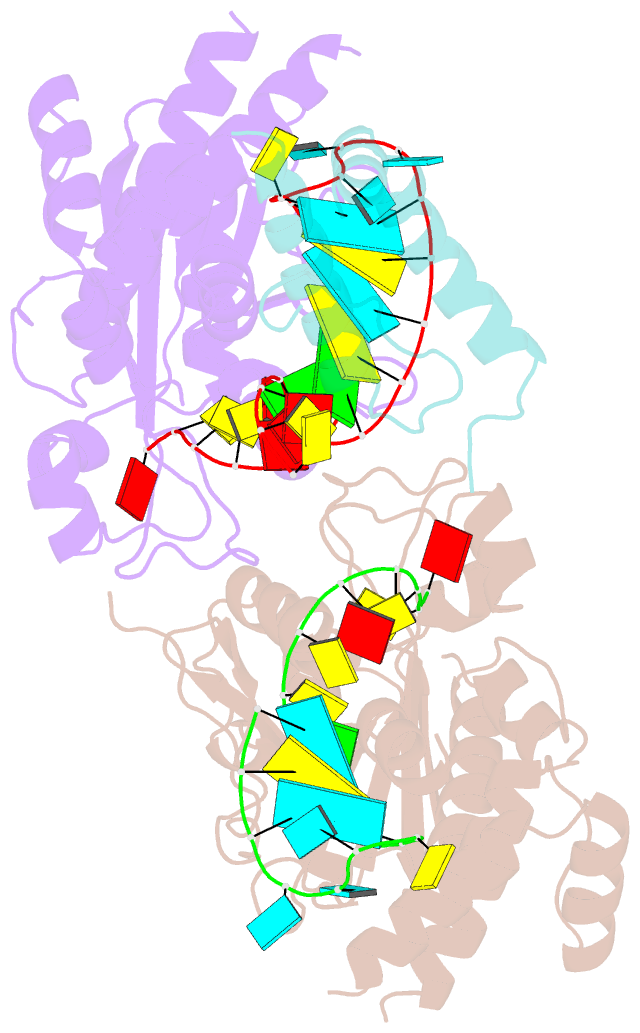
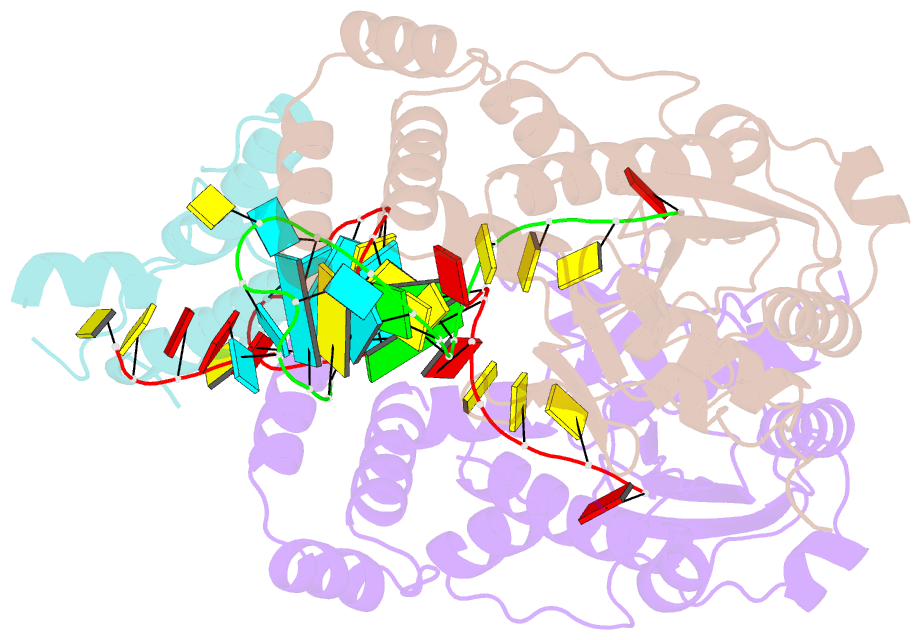
- Structure of mrna stem-loop, human stem-loop binding protein and 3'hexo ternary complex
- Reference
- Tan D, Marzluff WF, Dominski Z, Tong L (2013): "Structure of Histone Mrna Stem-Loop, Human Stem-Loop Binding Protein, and 3'Hexo Ternary Complex." Science, 339, 318. doi: 10.1126/SCIENCE.1228705.
- Abstract
- Metazoan replication-dependent histone messenger RNAs (mRNAs) have a conserved stem-loop (SL) at their 3'-end. The stem-loop binding protein (SLBP) specifically recognizes the SL to regulate histone mRNA metabolism, and the 3'-5' exonuclease 3'hExo trims its 3'-end after processing. We report the crystal structure of a ternary complex of human SLBP RNA binding domain, human 3'hExo, and a 26-nucleotide SL RNA. Only one base of the SL is recognized specifically by SLBP, and the two proteins primarily recognize the shape of the RNA. SLBP and 3'hExo have no direct contact with each other, and induced structural changes in the loop of the SL mediate their cooperative binding. The 3' flanking sequence is positioned in the 3'hExo active site, but the ternary complex limits the extent of trimming.