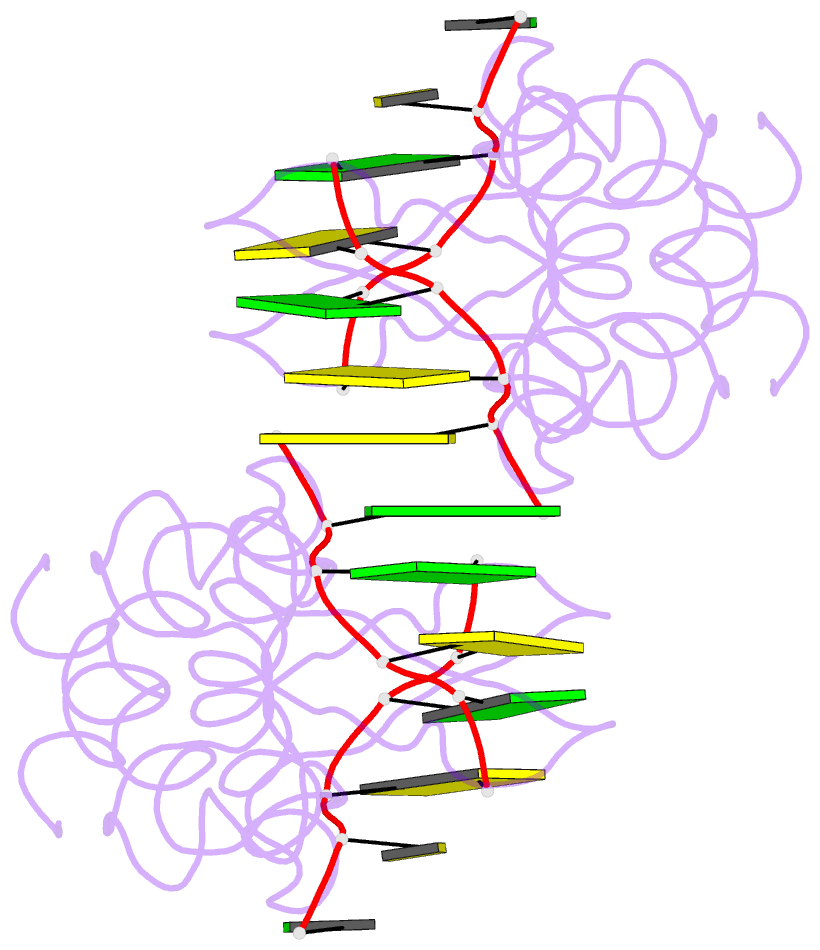
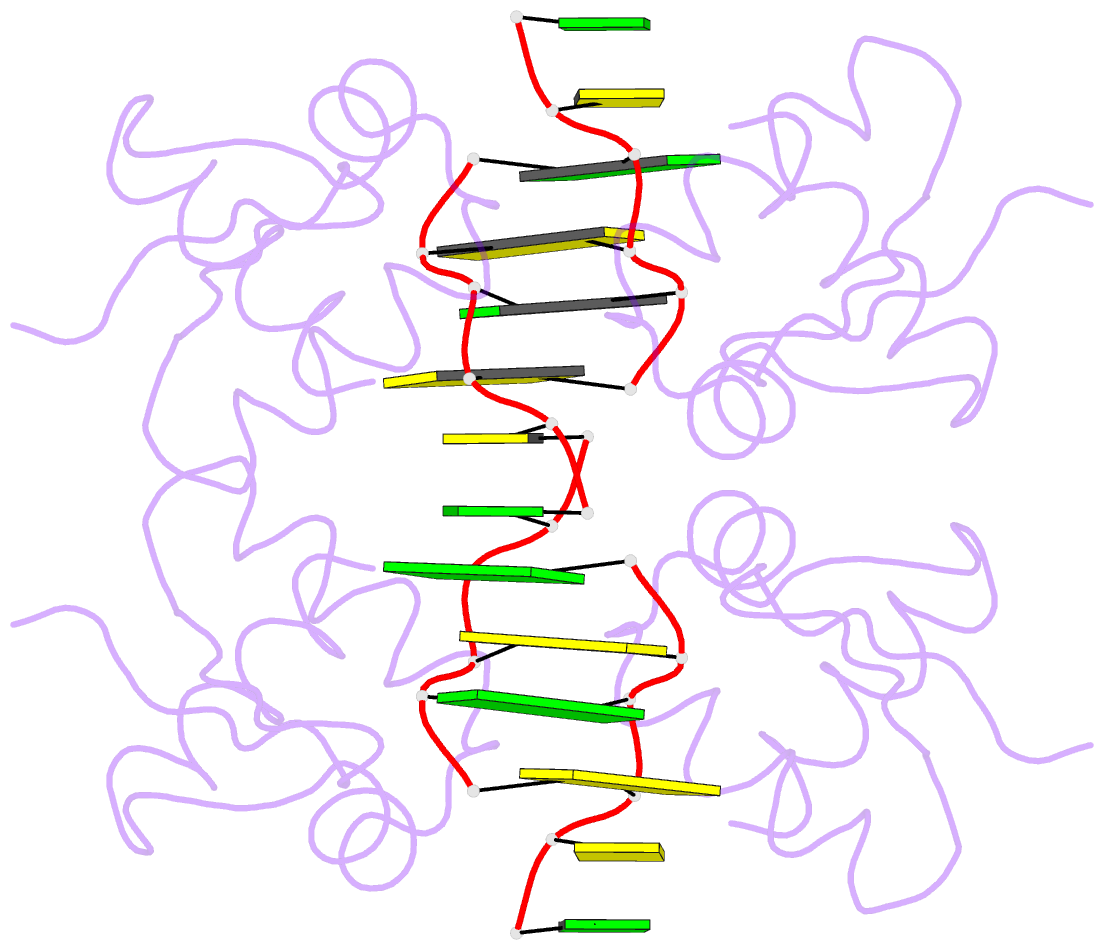
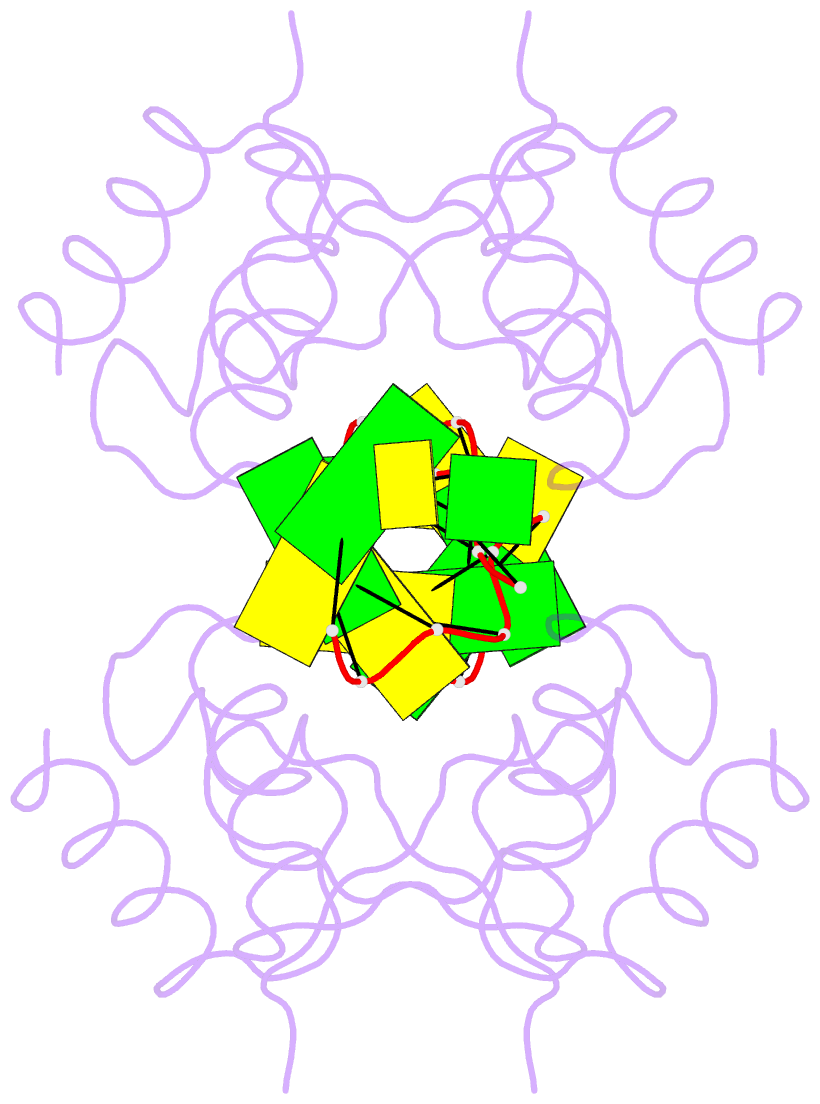
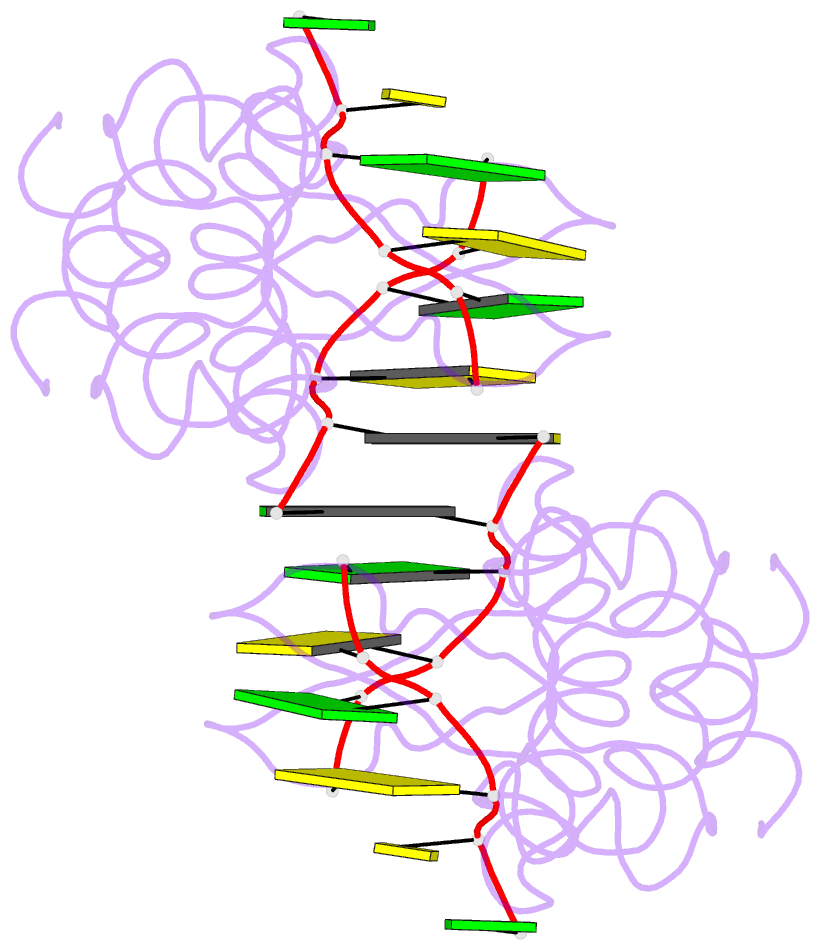
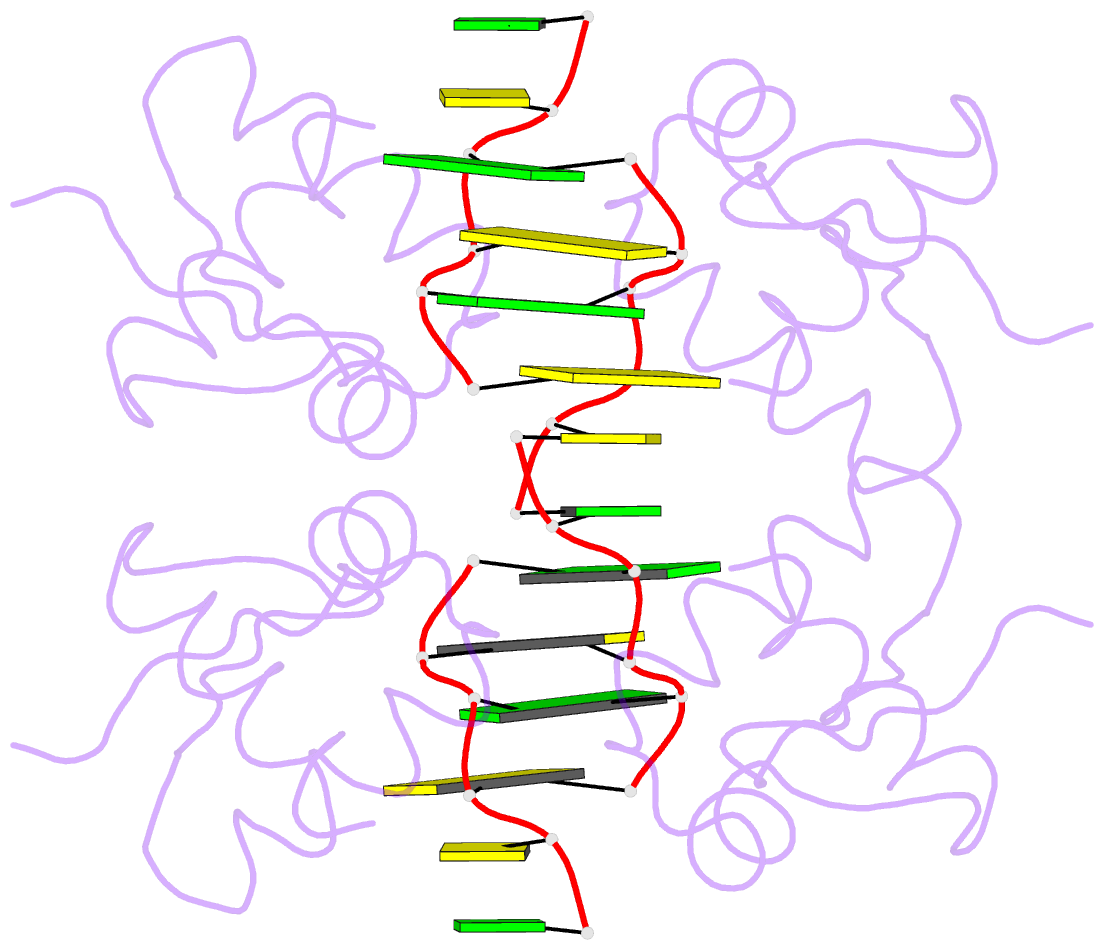
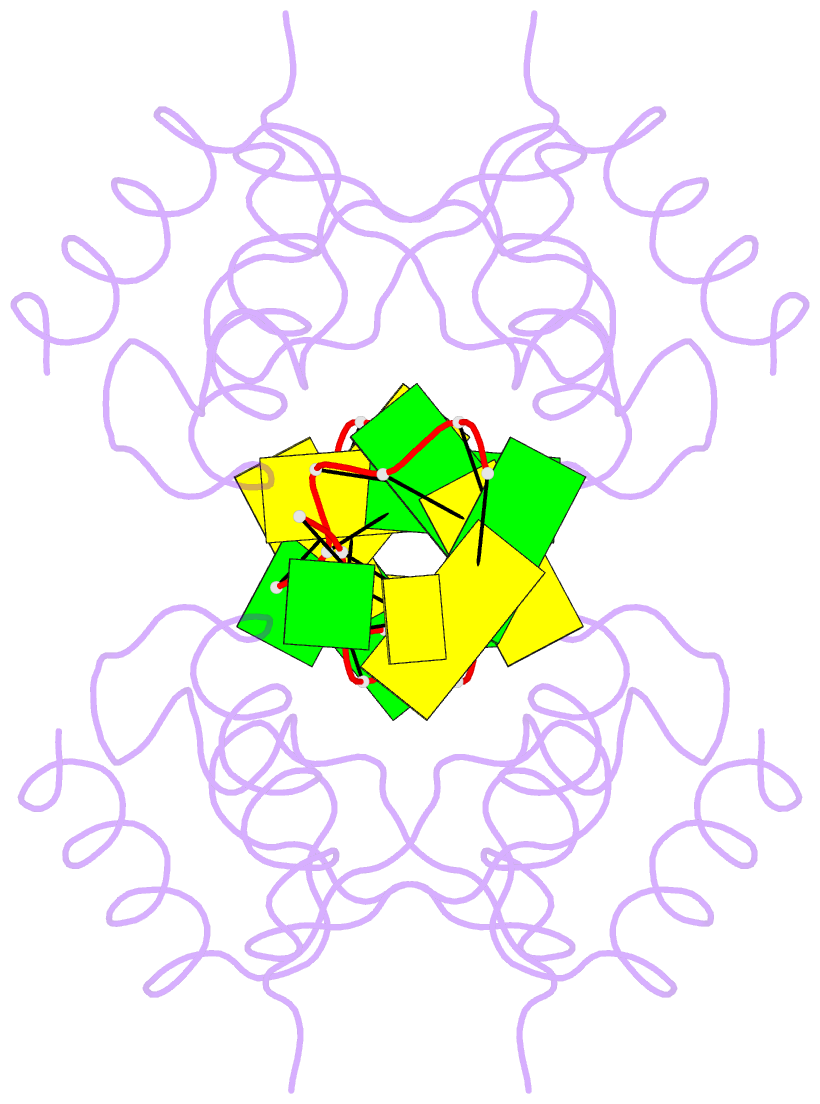
Summary information and primary citation
- PDB-id
- 4lb6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.8 Å)
- Summary
- Crystal structure of pkz zalpha in complex with ds(cg)6 (tetragonal form)
- Reference
- de Rosa M, Zacarias S, Athanasiadis A (2013): "Structural basis for Z-DNA binding and stabilization by the zebrafish Z-DNA dependent protein kinase PKZ." Nucleic Acids Res., 41, 9924-9933. doi: 10.1093/nar/gkt743.
- Abstract
- The RNA-dependent protein kinase PKR plays a central role in the antiviral defense of vertebrates by shutting down protein translation upon detection of viral dsRNA in the cytoplasm. In some teleost fish, PKZ, a homolog of PKR, performs the same function, but surprisingly, instead of dsRNA binding domains, it harbors two Z-DNA/Z-RNA-binding domains belonging to the Zalpha domain family. Zalpha domains have also been found in other proteins, which have key roles in the regulation of interferon responses such as ADAR1 and DNA-dependent activator of IFN-regulatory factors (DAI) and in viral proteins involved in immune response evasion such as the poxviral E3L and the Cyprinid Herpesvirus 3 ORF112. The underlying mechanism of nucleic acids binding and stabilization by Zalpha domains is still unclear. Here, we present two crystal structures of the zebrafish PKZ Zalpha domain (DrZalpha(PKZ)) in alternatively organized complexes with a (CG)6 DNA oligonucleotide at 2 and 1.8 Å resolution. These structures reveal novel aspects of the Zalpha interaction with DNA, and they give insights on the arrangement of multiple Zalpha domains on DNA helices longer than the minimal binding site.