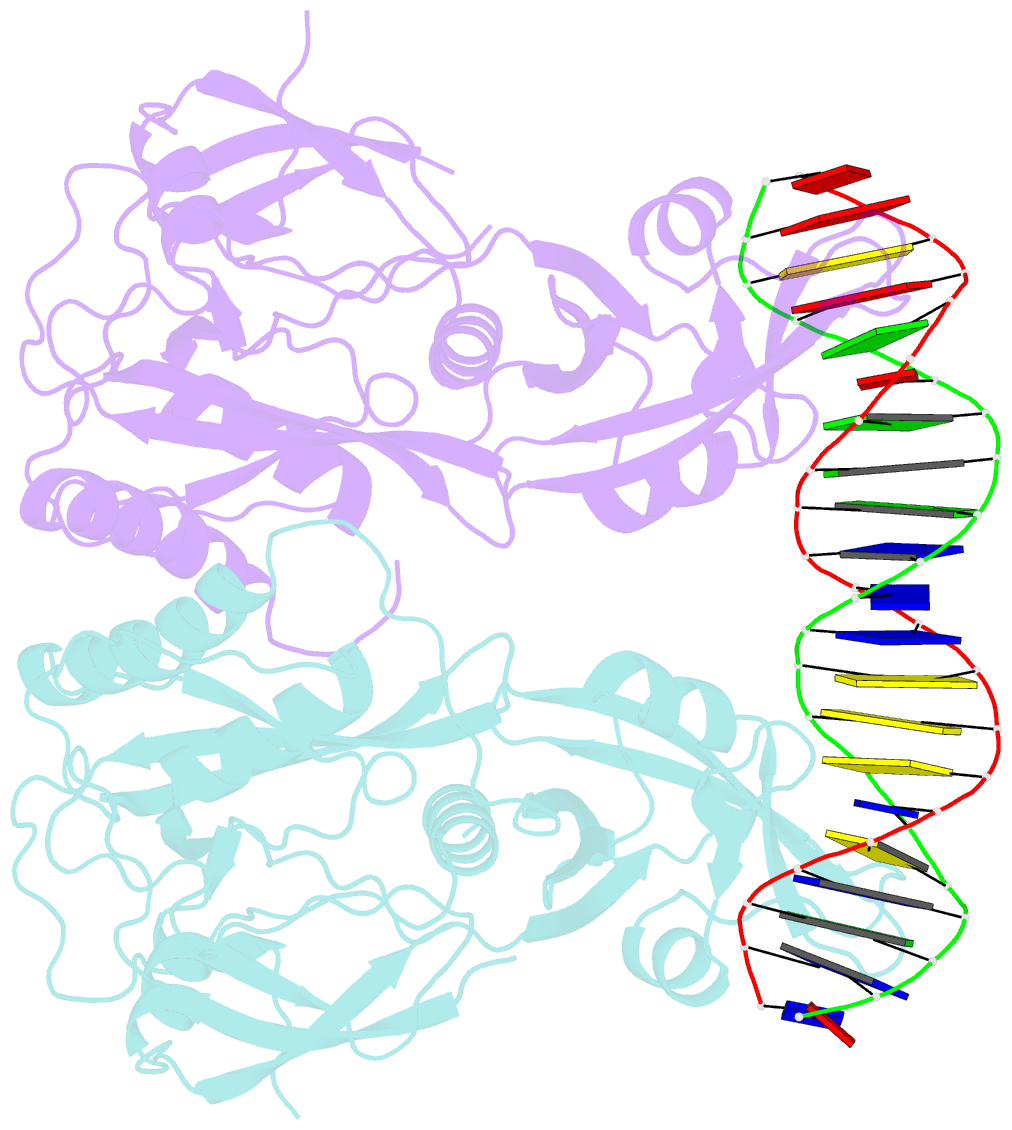
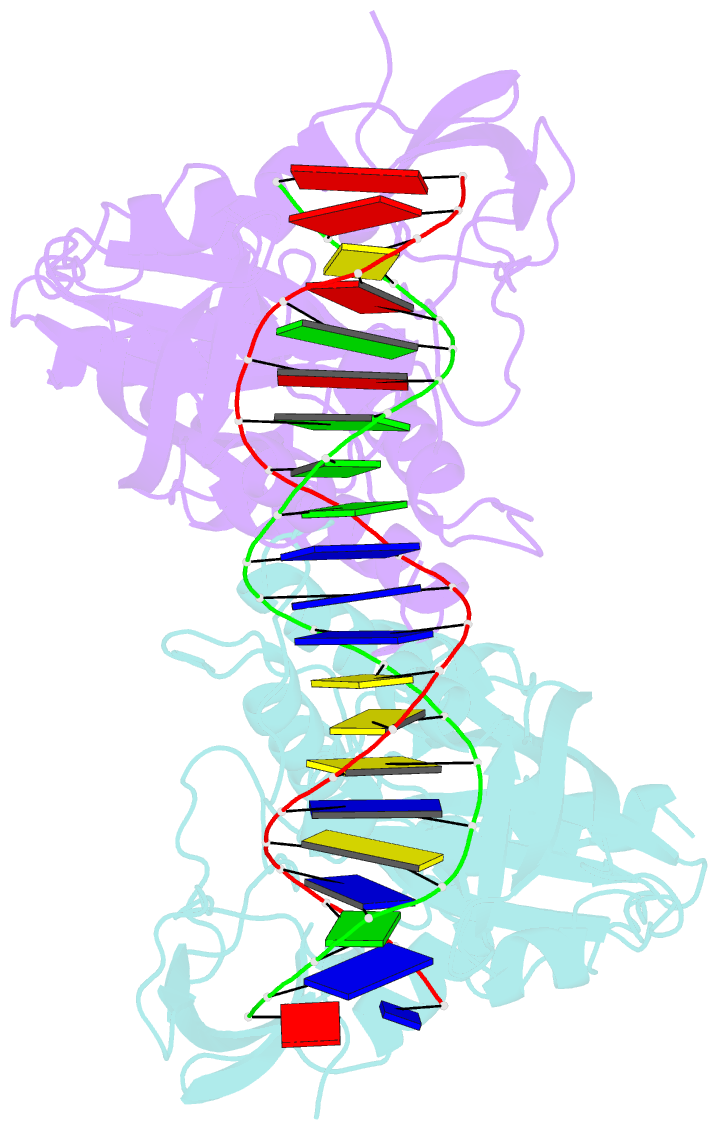
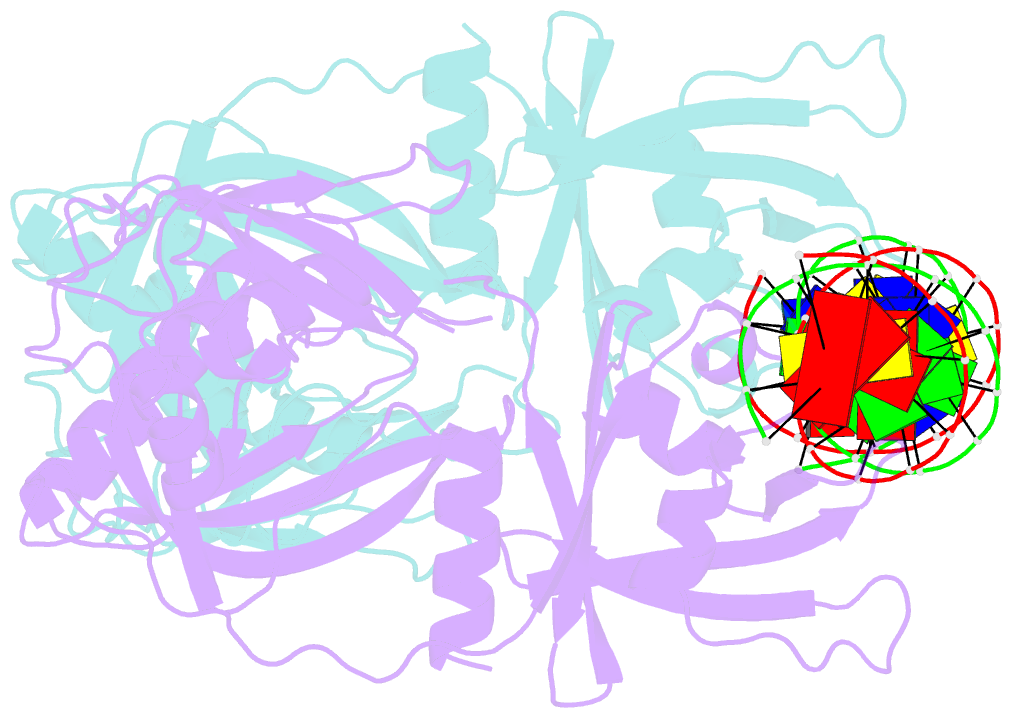
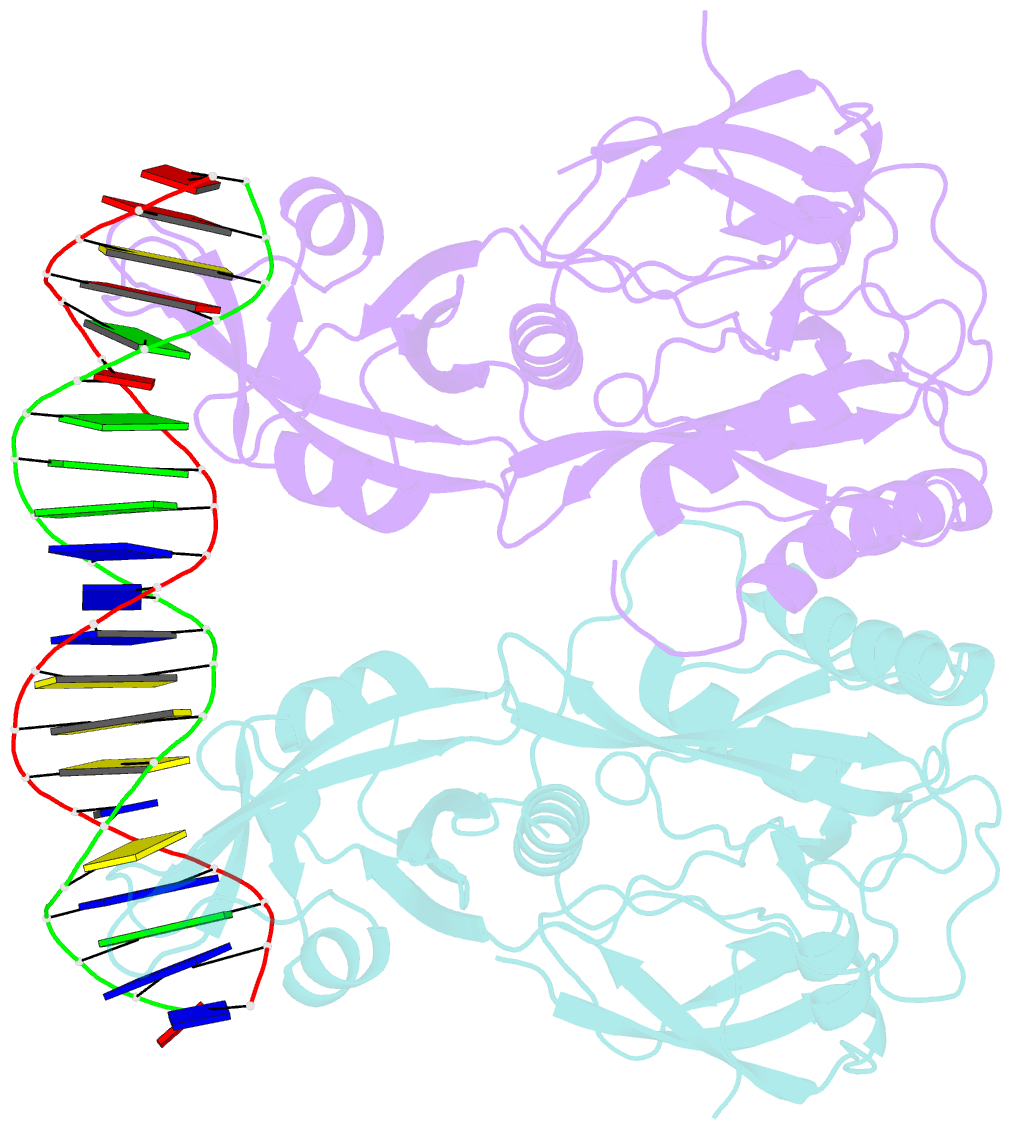
Summary information and primary citation
- PDB-id
- 4ldx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.9 Å)
- Summary
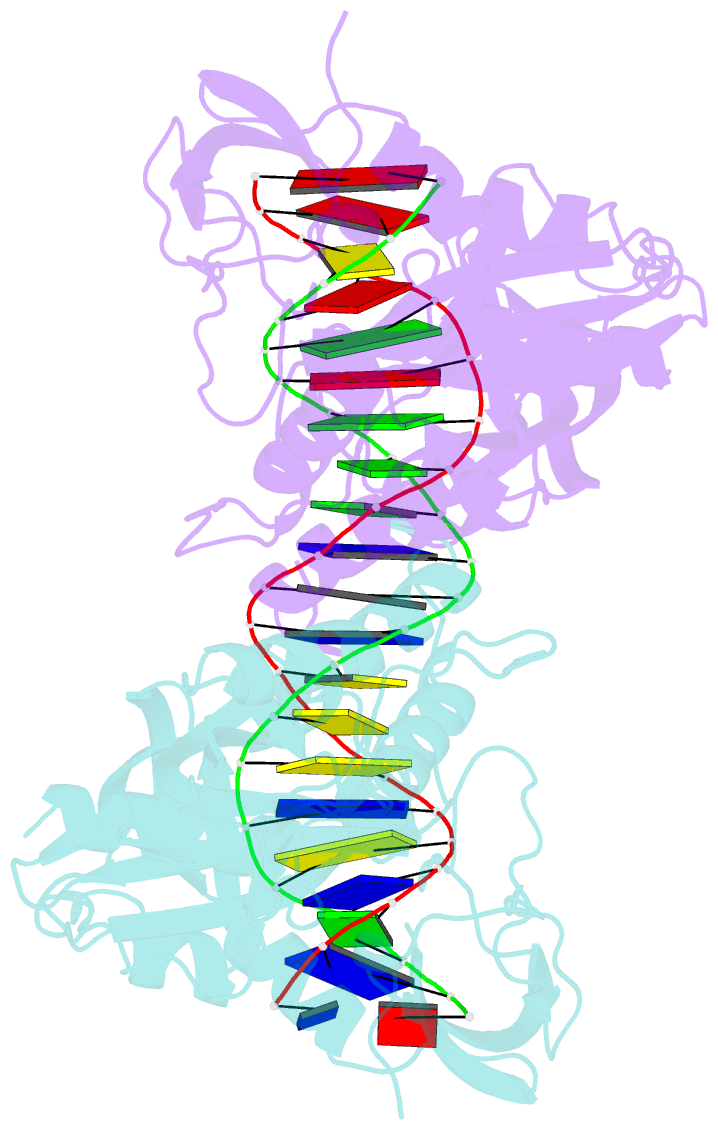
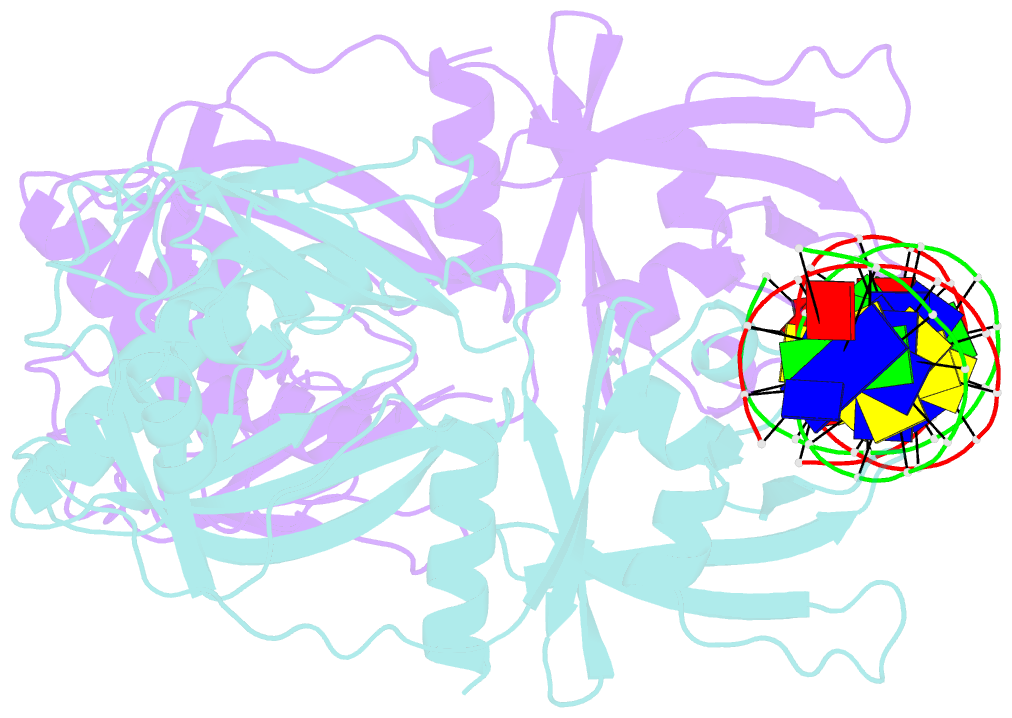
- Crystal structure of the DNA binding domain of arabidopsis thaliana auxin response factor 1 (arf1) in complex with protomor-like sequence er7
- Reference
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, Lopez-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, Weijers D, Coll M (2014): "Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors." Cell(Cambridge,Mass.), 156, 577-589. doi: 10.1016/j.cell.2013.12.027.
- Abstract
- Auxin regulates numerous plant developmental processes by controlling gene expression via a family of functionally distinct DNA-binding auxin response factors (ARFs), yet the mechanistic basis for generating specificity in auxin response is unknown. Here, we address this question by solving high-resolution crystal structures of the pivotal Arabidopsis developmental regulator ARF5/MONOPTEROS (MP), its divergent paralog ARF1, and a complex of ARF1 and a generic auxin response DNA element (AuxRE). We show that ARF DNA-binding domains also homodimerize to generate cooperative DNA binding, which is critical for in vivo ARF5/MP function. Strikingly, DNA-contacting residues are conserved between ARFs, and we discover that monomers have the same intrinsic specificity. ARF1 and ARF5 homodimers, however, differ in spacing tolerated between binding sites. Our data identify the DNA-binding domain as an ARF dimerization domain, suggest that ARF dimers bind complex sites as molecular calipers with ARF-specific spacing preference, and provide an atomic-scale mechanistic model for specificity in auxin response.