Summary information and primary citation
- PDB-id
- 4lgt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-RNA
- Method
- X-ray (1.3 Å)
- Summary
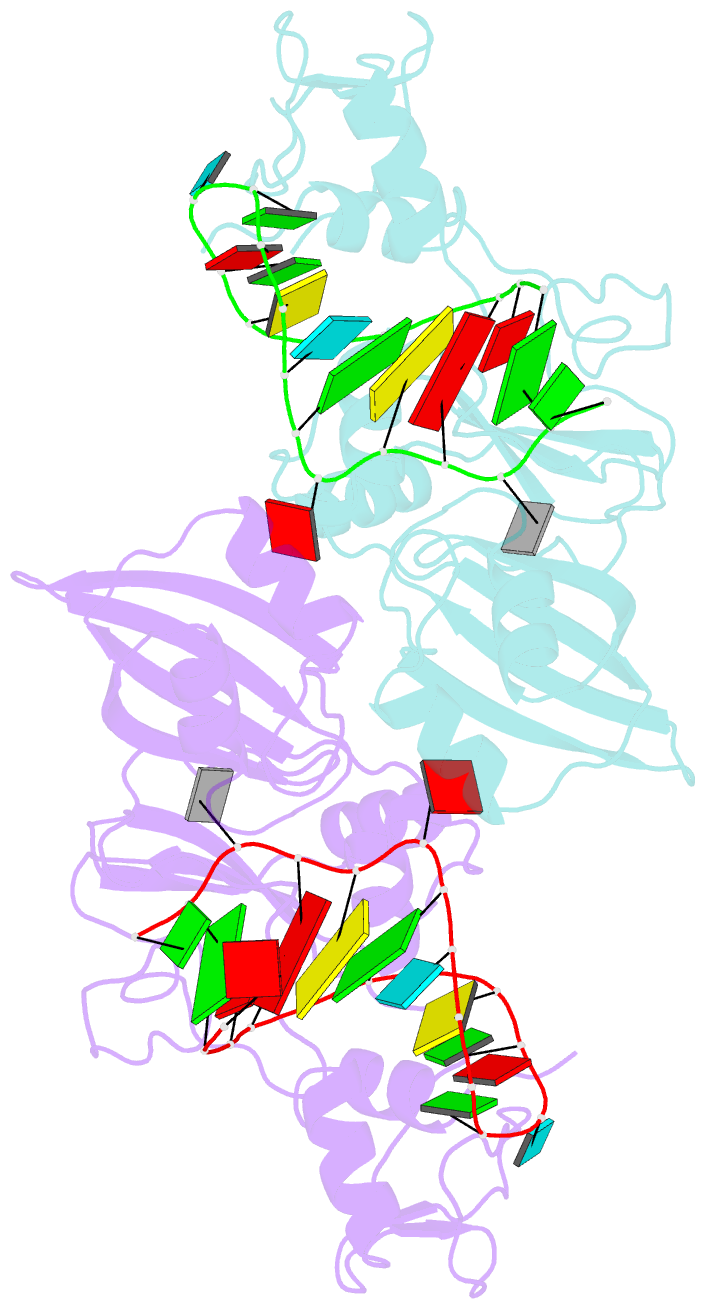
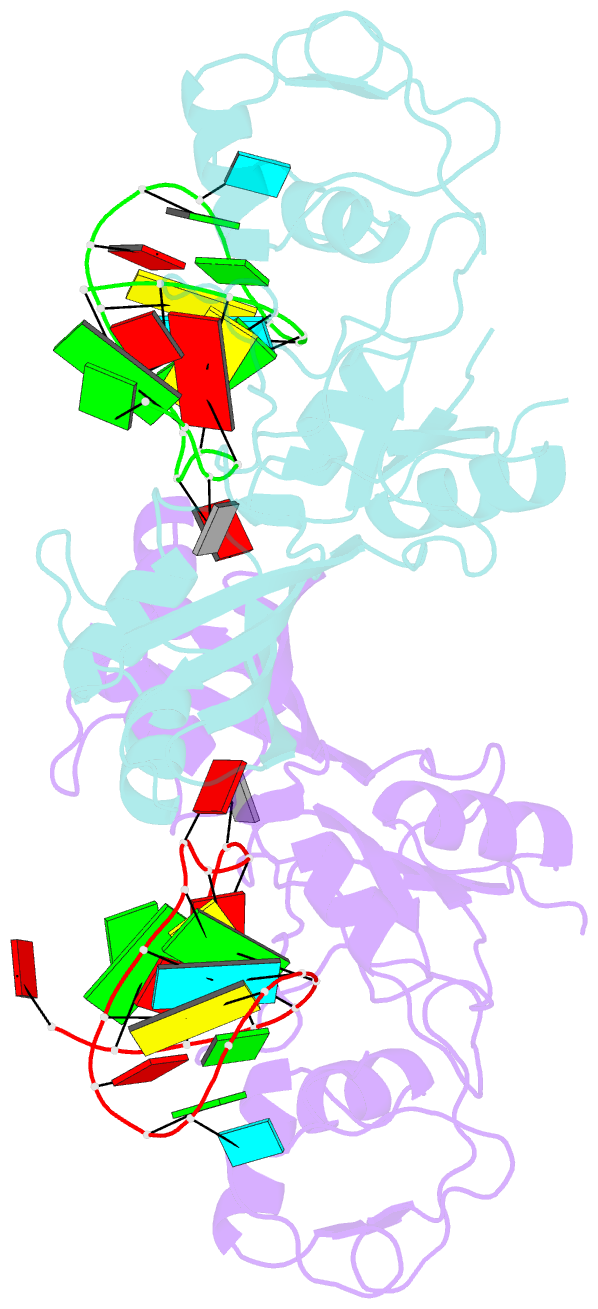
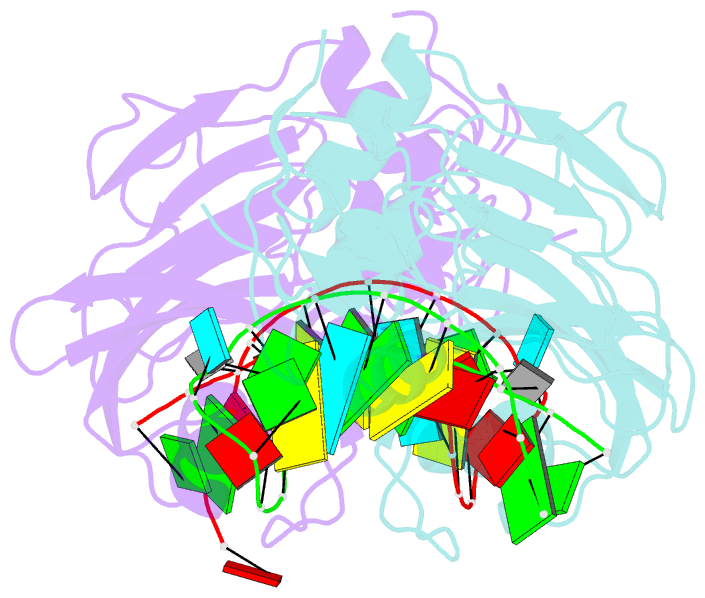
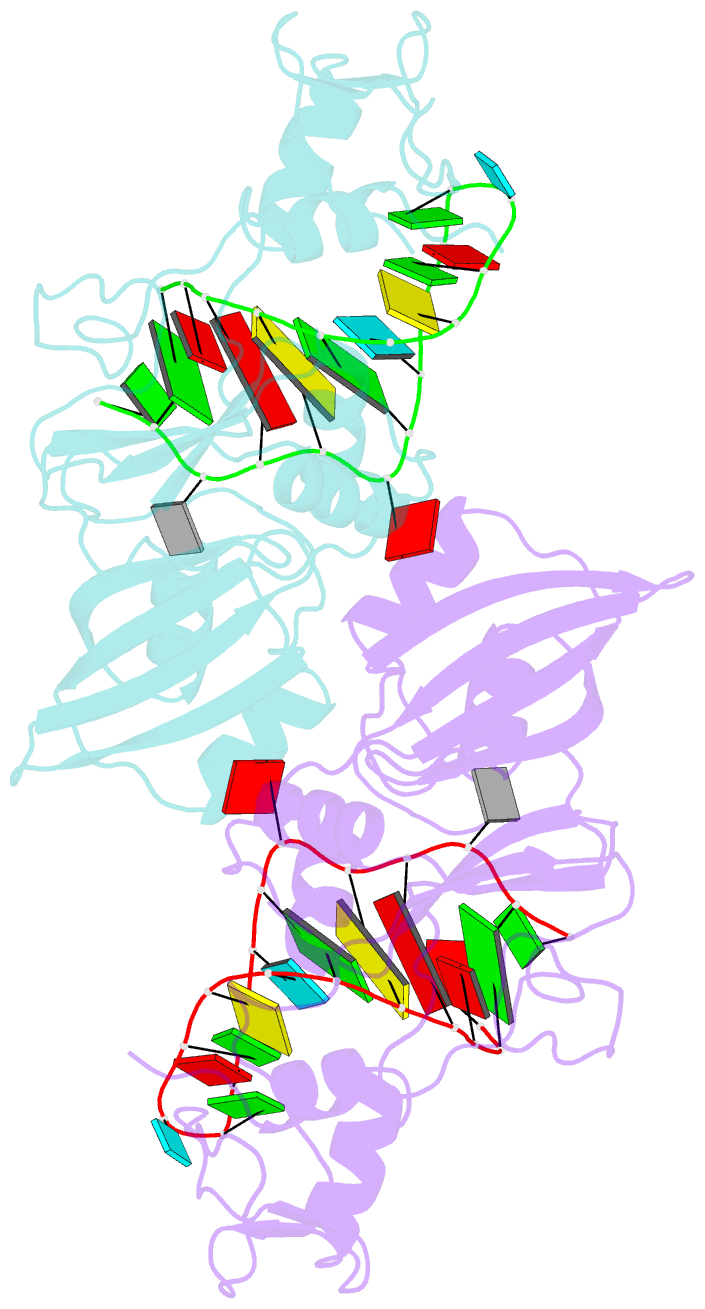
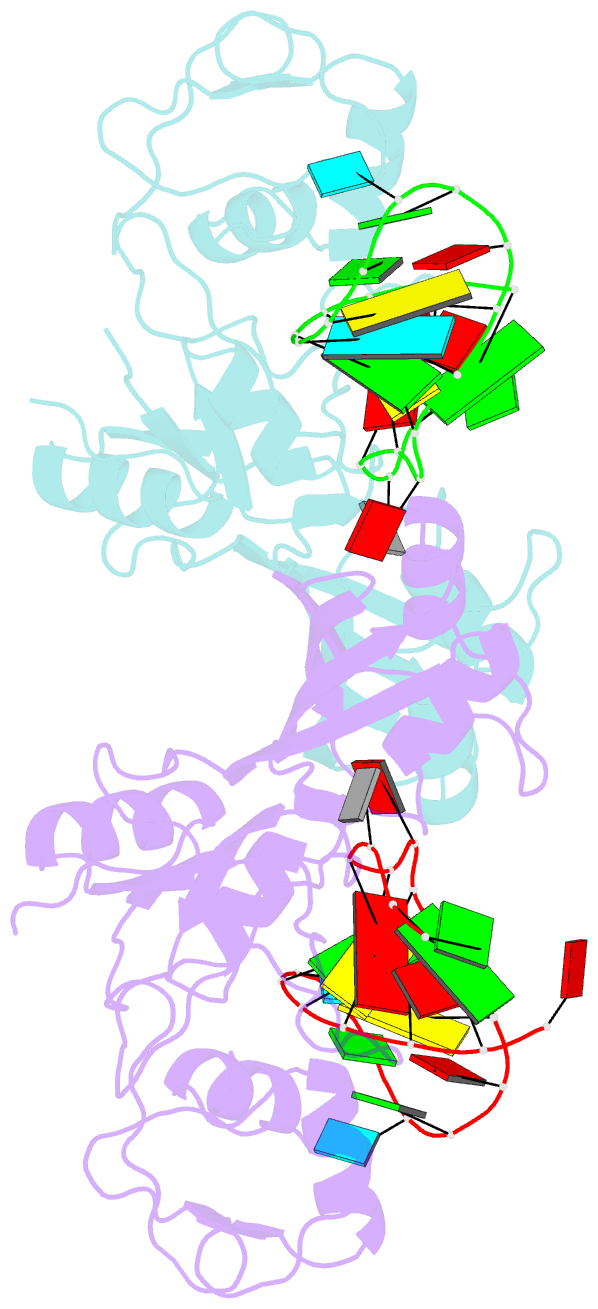
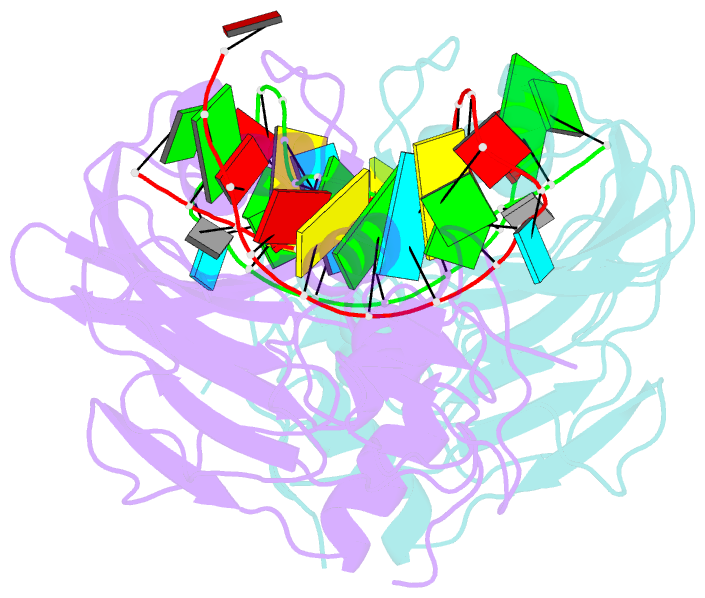
- Crystal structure of the catalytic domain of rlub in complex with a 21-nucleotide RNA substrate
- Reference
- Czudnochowski N, Ashley GW, Santi DV, Alian A, Finer-Moore J, Stroud RM (2014): "The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs." Nucleic Acids Res., 42, 2037-2048. doi: 10.1093/nar/gkt1050.
- Abstract
- RluB catalyses the modification of U2605 to pseudouridine (Ψ) in a stem-loop at the peptidyl transferase center of Escherichia coli 23S rRNA. The homolog RluF is specific to the adjacent nucleotide in the stem, U2604. The 1.3 Å resolution crystal structure of the complex between the catalytic domain of RluB and the isolated substrate stem-loop, in which the target uridine is substituted by 5-fluorouridine (5-FU), reveals a covalent bond between the isomerized target base and tyrosine 140. The structure is compared with the catalytic domain alone determined at 2.5 Å resolution. The RluB-bound stem-loop has essentially the same secondary structure as in the ribosome, with a bulge at A2602, but with 5-FU2605 flipped into the active site. We showed earlier that RluF induced a frame-shift of the RNA, moving A2602 into the stem and translating its target, U2604, into the active site. A hydrogen-bonding network stabilizes the bulge in the RluB-RNA but is not conserved in RluF and so RluF cannot stabilize the bulge. On the basis of the covalent bond between enzyme and isomerized 5-FU we propose a Michael addition mechanism for pseudouridine formation that is consistent with all experimental data.