Summary information and primary citation
- PDB-id
- 4lj0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein
- Method
- X-ray (2.15 Å)
- Summary
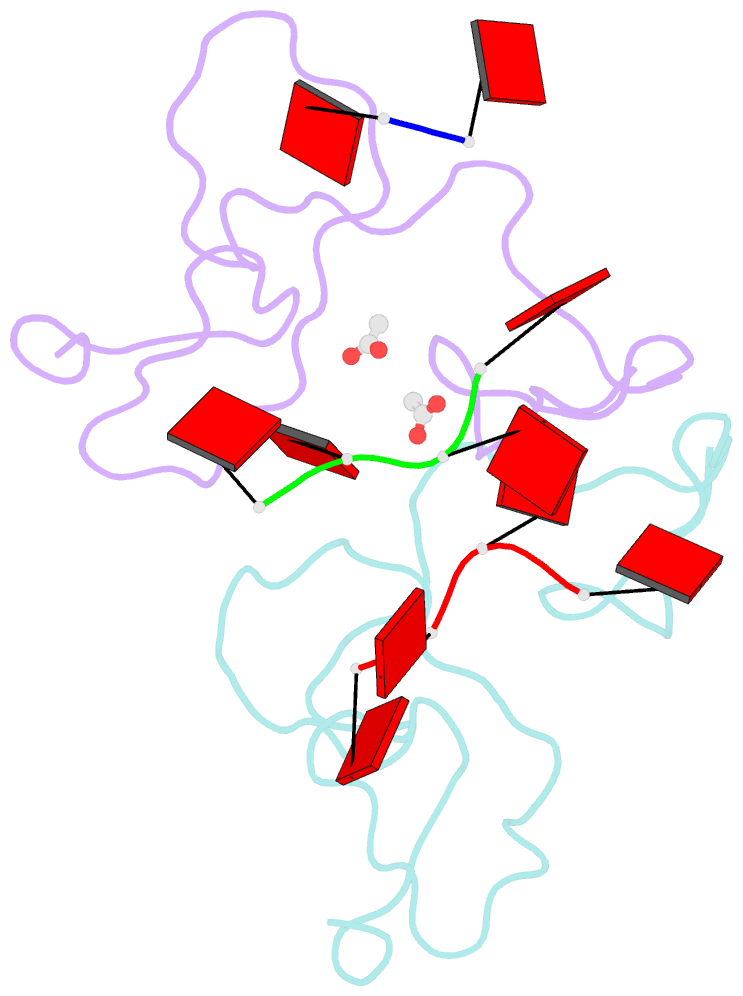
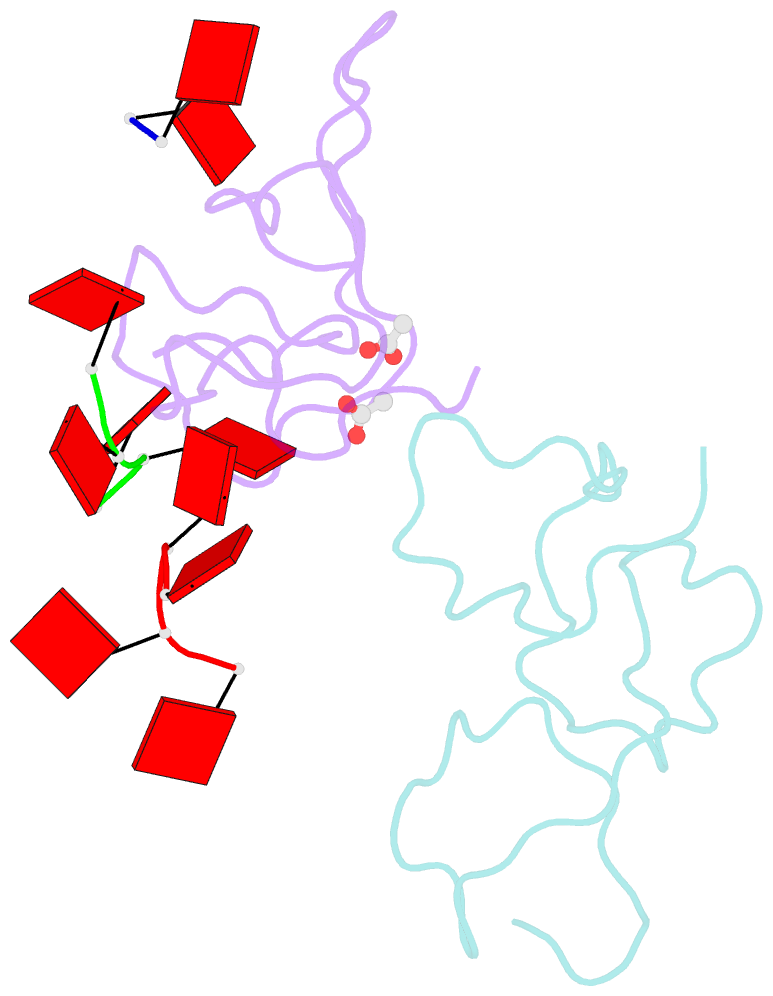
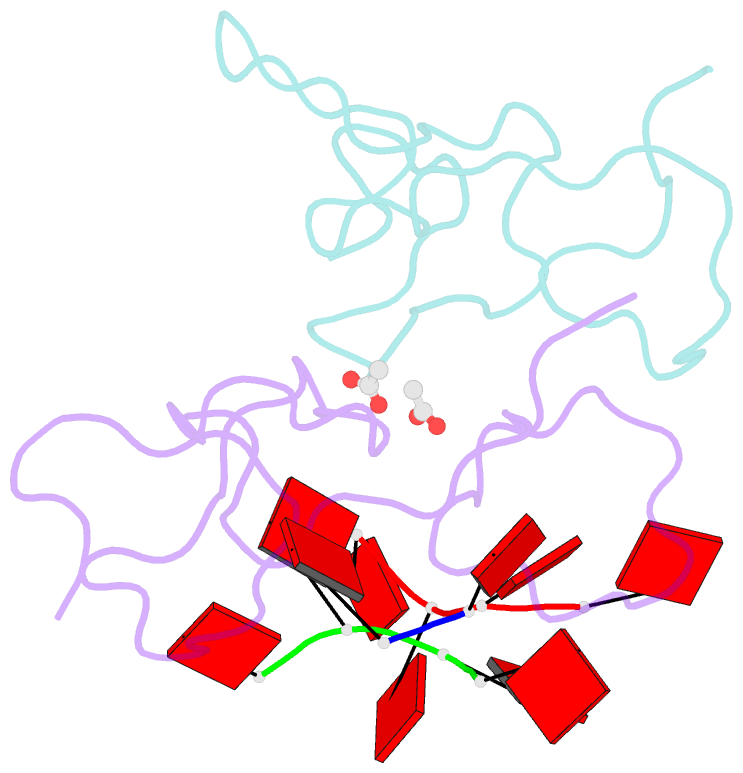
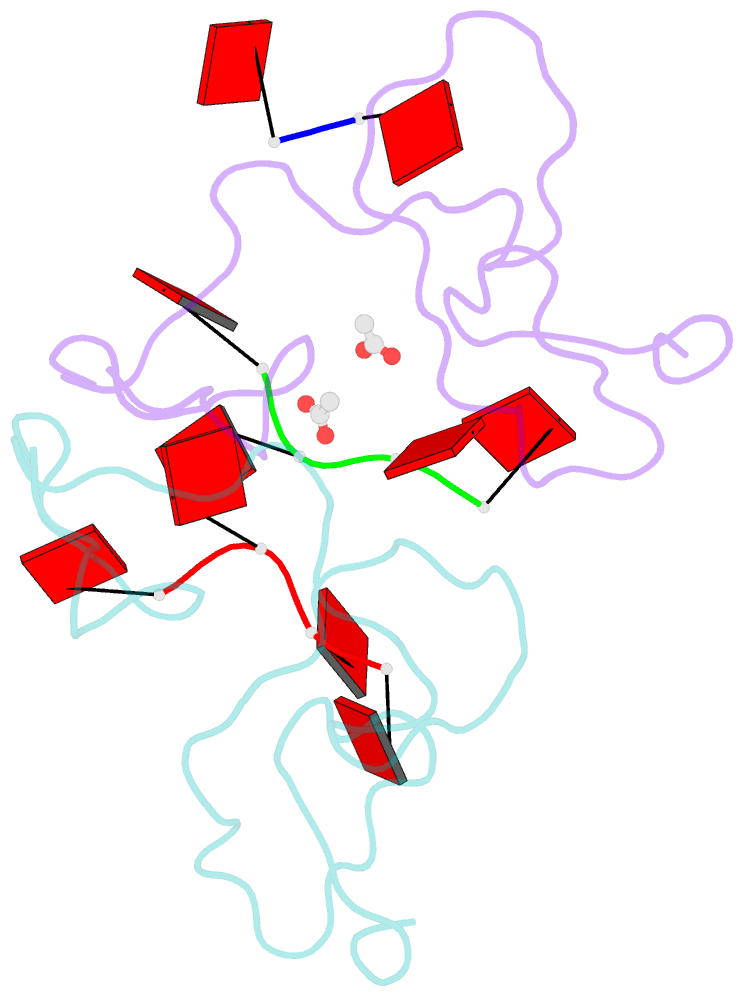
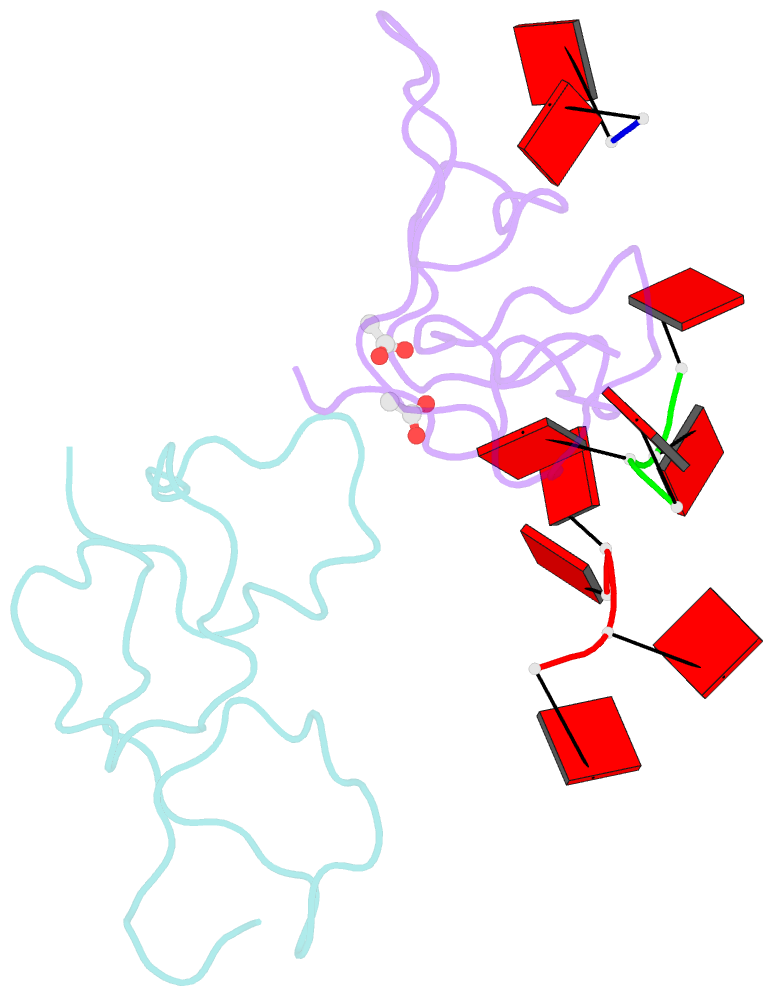
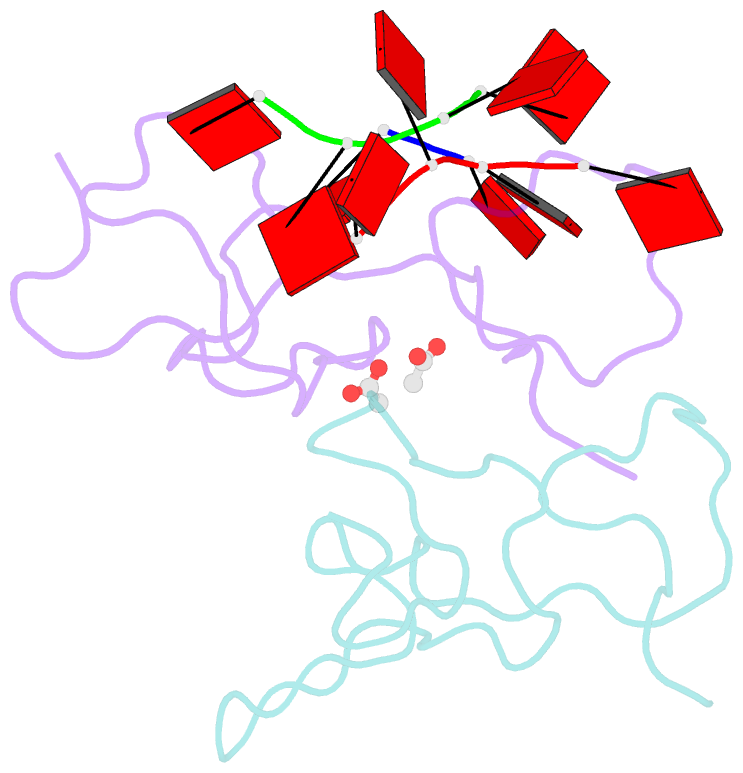
- Nab2 zn fingers complexed with polyadenosine
- Reference
- Kuhlmann SI, Valkov E, Stewart M (2014): "Structural basis for the molecular recognition of polyadenosine RNA by Nab2 Zn fingers." Nucleic Acids Res., 42, 672-680. doi: 10.1093/nar/gkt876.
- Abstract
- The yeast poly(A) RNA binding protein, Nab2, facilitates poly(A) tail length regulation together with targeting transcripts to nuclear pores and their export to the cytoplasm. Nab2 binds polyadenosine RNA primarily through a tandem repeat of CCCH Zn fingers. We report here the 2.15 Å resolution crystal structure of Zn fingers 3-5 of Chaetomium thermophilum Nab2 bound to polyadenosine RNA and establish the structural basis for the molecular recognition of adenosine ribonucleotides. Zn fingers 3 and 5 each bind two adenines, whereas finger 4 binds only one. In each case, the purine ring binds in a surface groove, where it stacks against an aromatic side chain, with specificity being provided by a novel pattern of H-bonds, most commonly between purine N6 and a Zn-coordinated cysteine supplemented by H-bonds between purine N7 and backbone amides. Residues critical for adenine binding are conserved between species and provide a code that allows prediction of finger-binding stoichiometry based on their sequence. Moreover, these results indicate that, in addition to poly(A) tails, Nab2 can also recognize sequence motifs elsewhere in transcripts in which adenosines are placed at key positions, consistent with its function in mRNP organization and compaction as well as poly(A) tail length regulation.