Summary information and primary citation
- PDB-id
- 4m94; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.14 Å)
- Summary
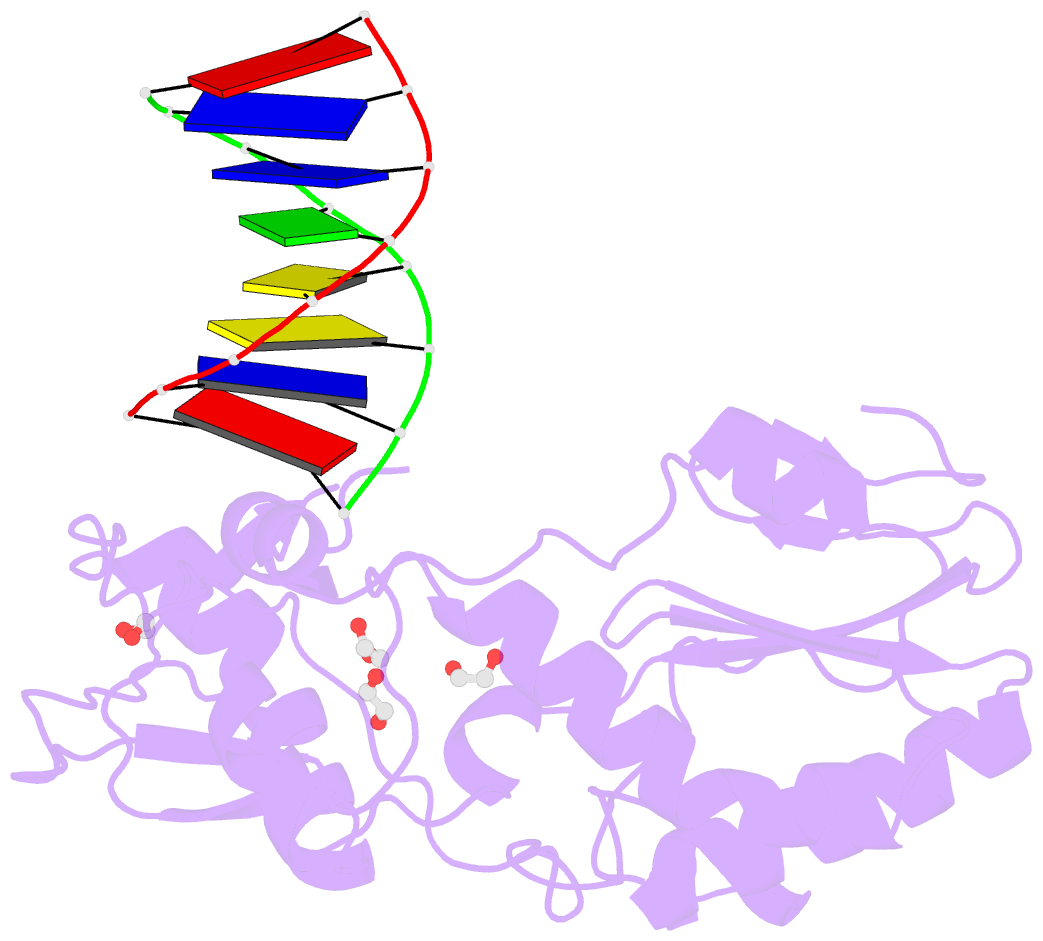
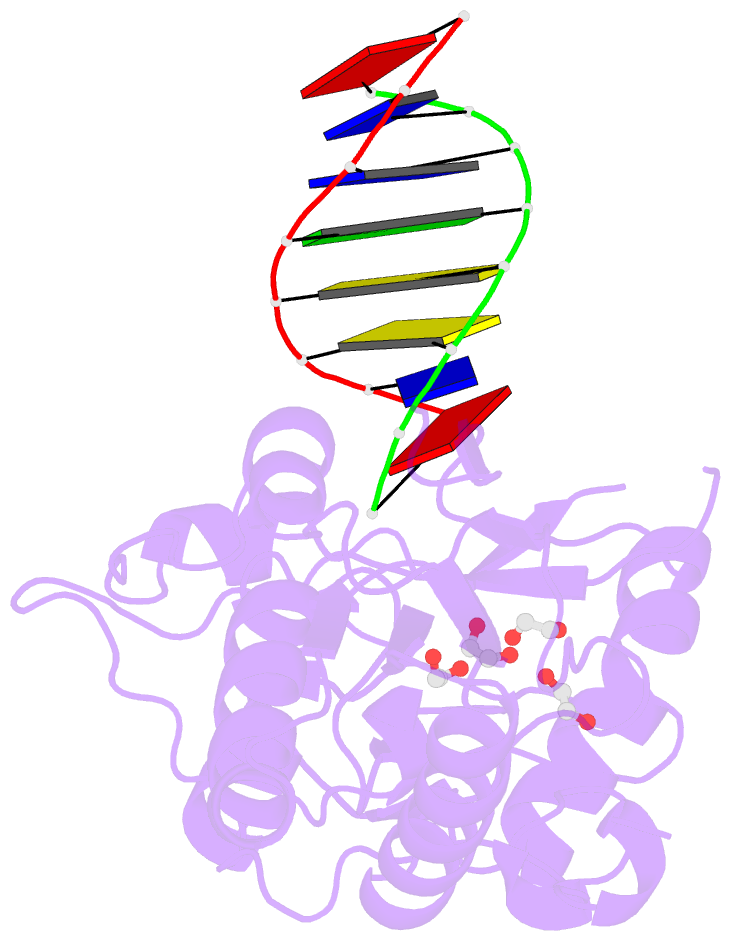
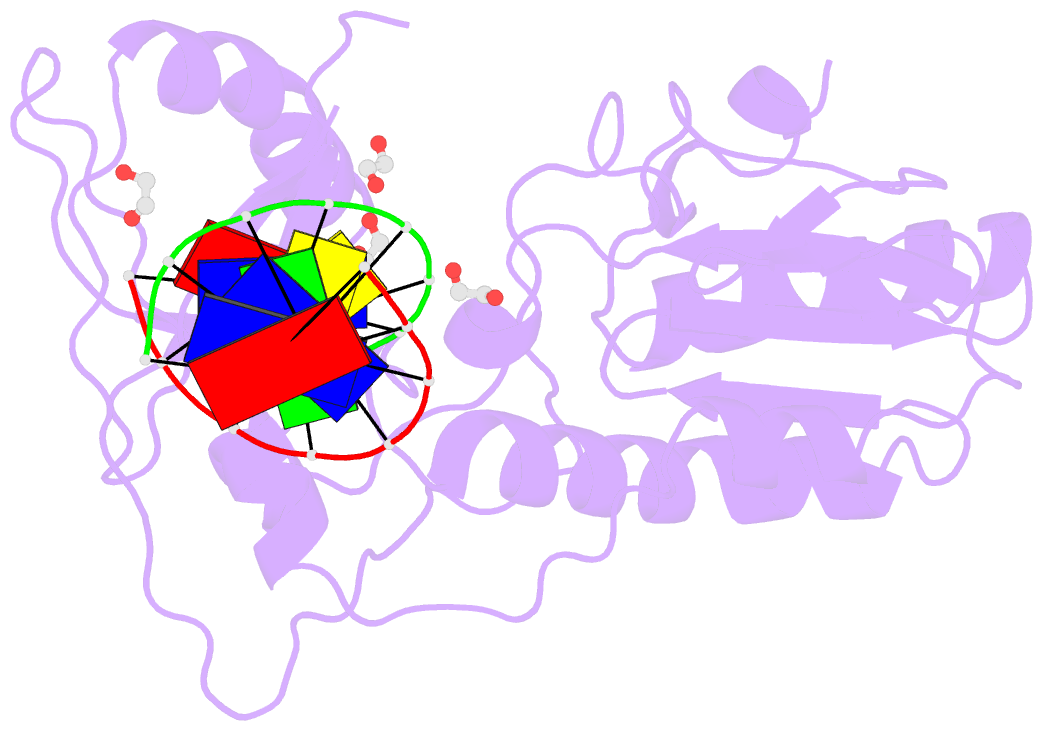
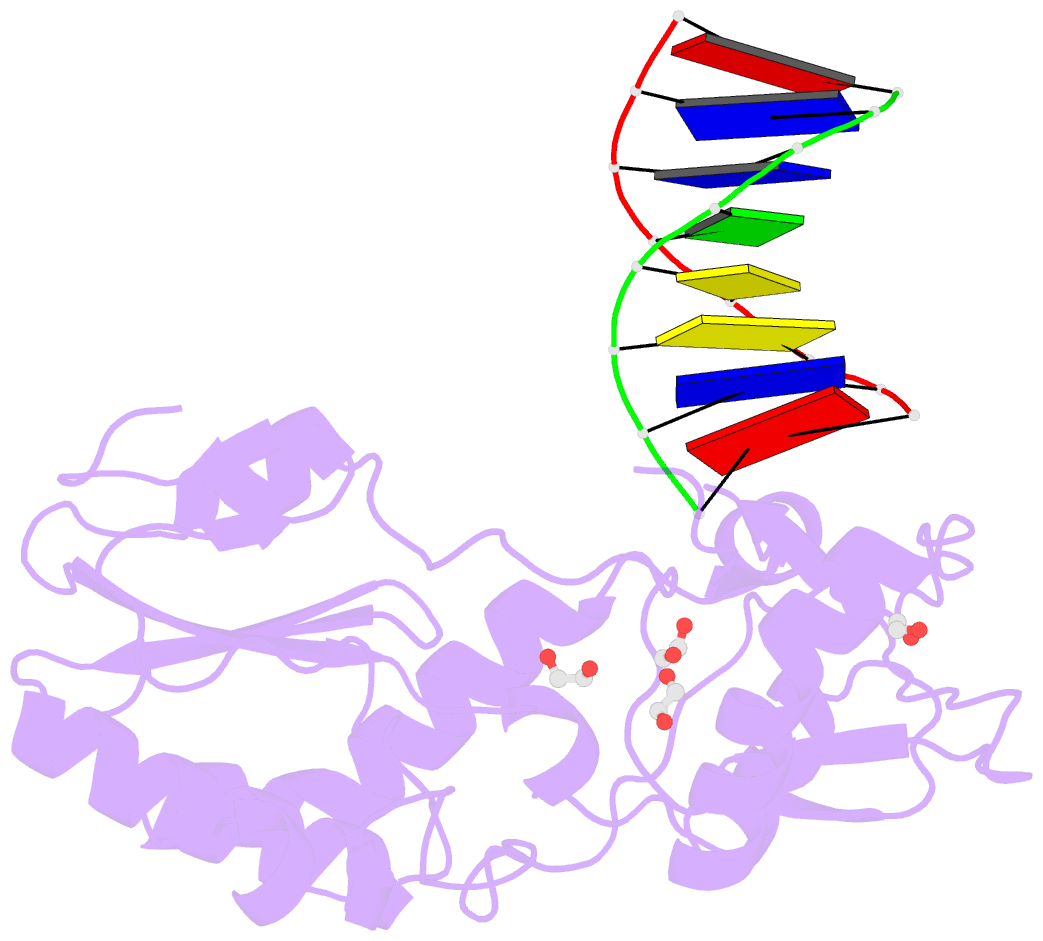
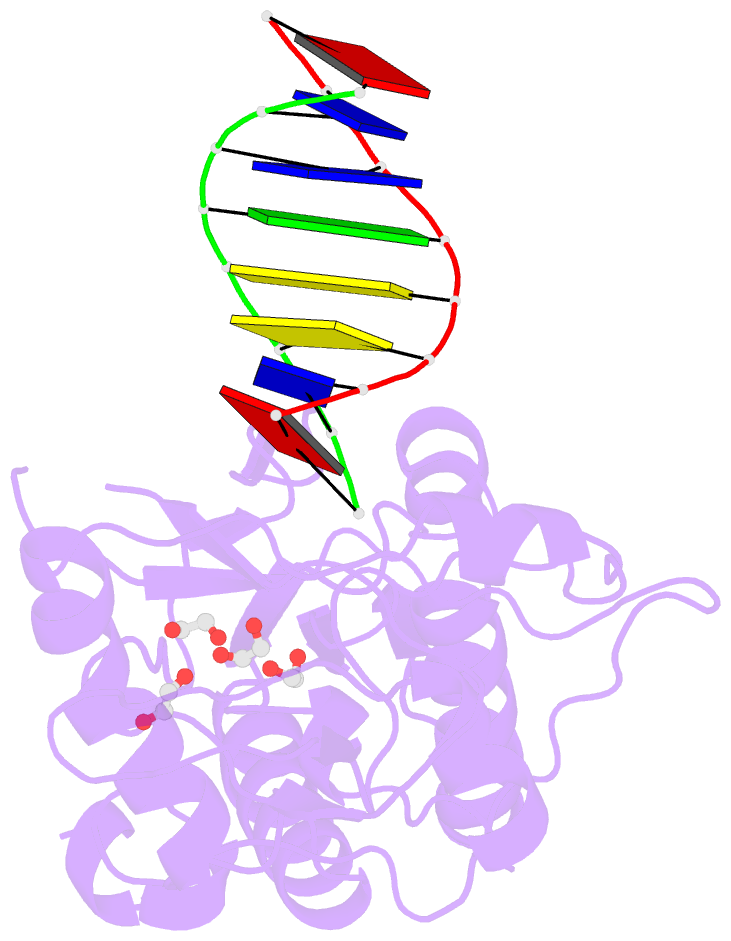
- D(atccgttataacggat) complexed with moloney murine leukemia virus reverse transcriptase catalytic fragment
- Reference
- Singh I, Lian Y, Li L, Georgiadis MM (2014): "The structure of an authentic spore photoproduct lesion in DNA suggests a basis for recognition." Acta Crystallogr.,Sect.D, 70, 752-759. doi: 10.1107/S1399004713032987.
- Abstract
- The spore photoproduct lesion (SP; 5-thymine-5,6-dihydrothymine) is the dominant photoproduct found in UV-irradiated spores of some bacteria such as Bacillus subtilis. Upon spore germination, this lesion is repaired in a light-independent manner by a specific repair enzyme: the spore photoproduct lyase (SP lyase). In this work, a host-guest approach in which the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase (MMLV RT) serves as the host and DNA as the guest was used to determine the crystal structures of complexes including 16 bp oligonucleotides with and without the SP lesion at 2.14 and 1.72 Å resolution, respectively. In contrast to other types of thymine-thymine lesions, the SP lesion retains normal Watson-Crick hydrogen bonding to the adenine bases of the complementary strand, with shorter hydrogen bonds than found in the structure of the undamaged DNA. However, the lesion induces structural changes in the local conformation of what is otherwise B-form DNA. The region surrounding the lesion differs significantly in helical form from B-DNA, and the minor groove is widened by almost 3 Å compared with that of the undamaged DNA. Thus, these unusual structural features associated with SP lesions may provide a basis for recognition by the SP lyase.