Summary information and primary citation
- PDB-id
- 4nid; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- oxidoreductase-DNA
- Method
- X-ray (1.58 Å)
- Summary
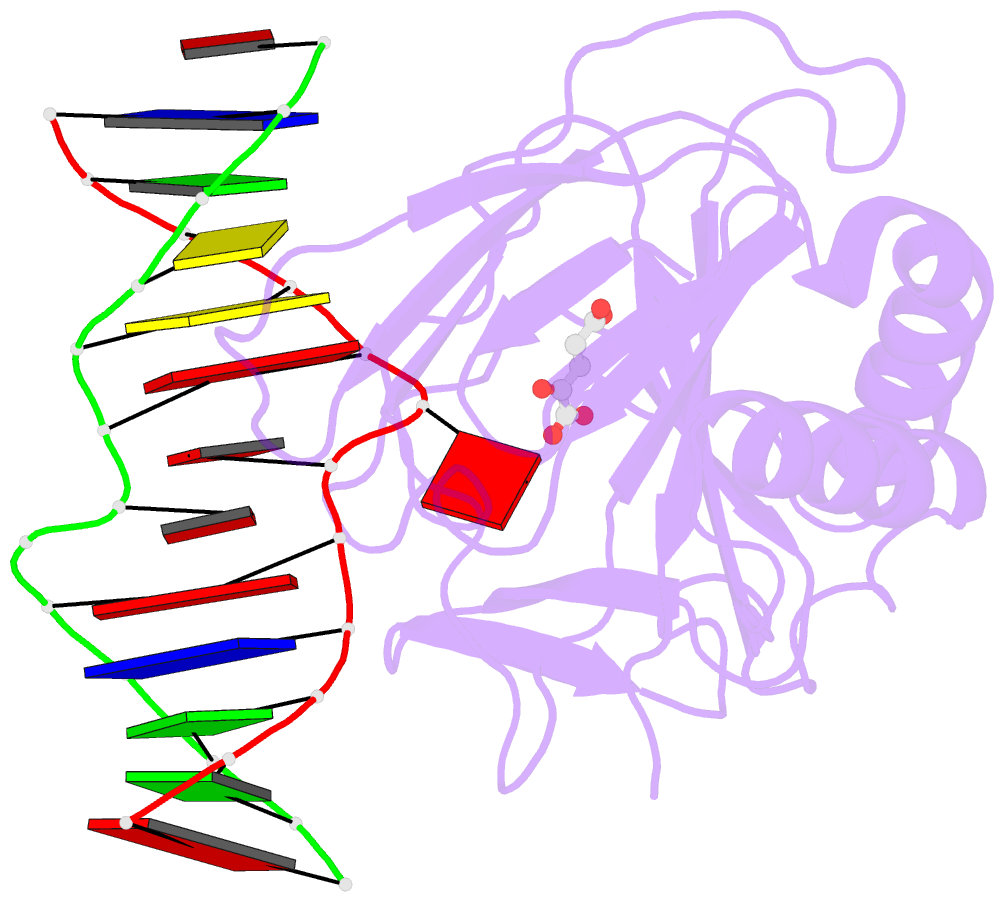
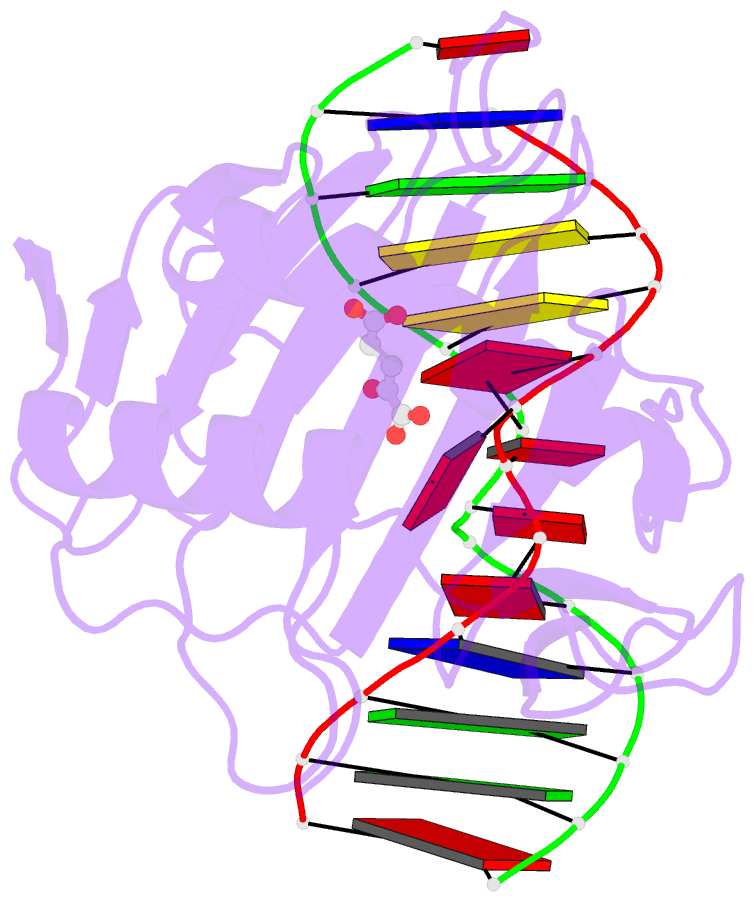
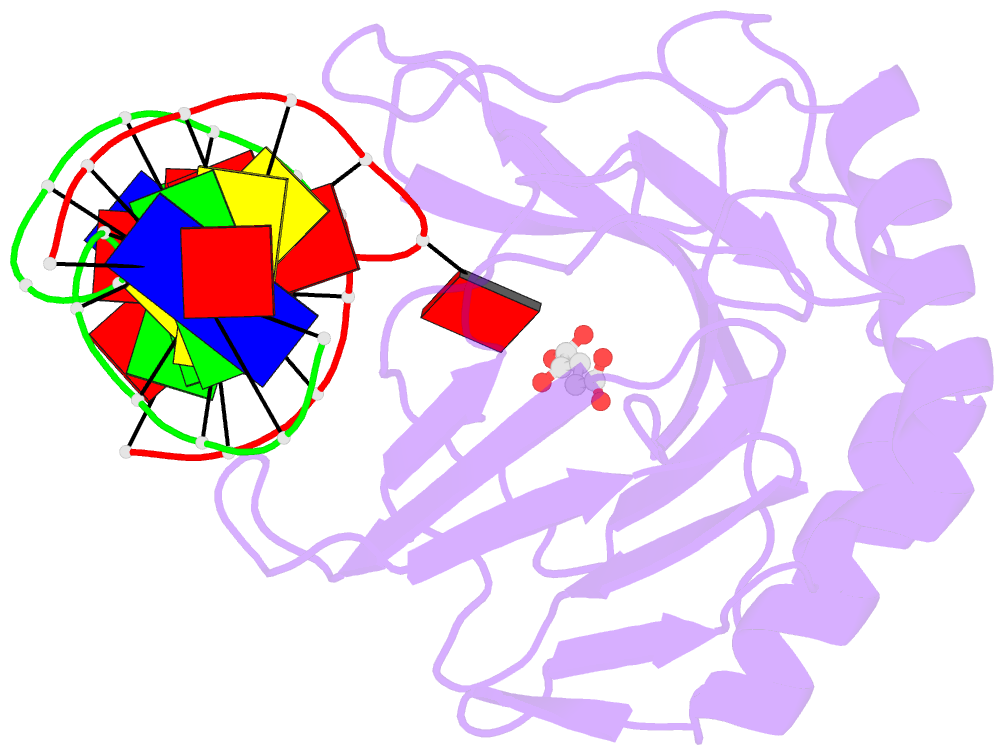
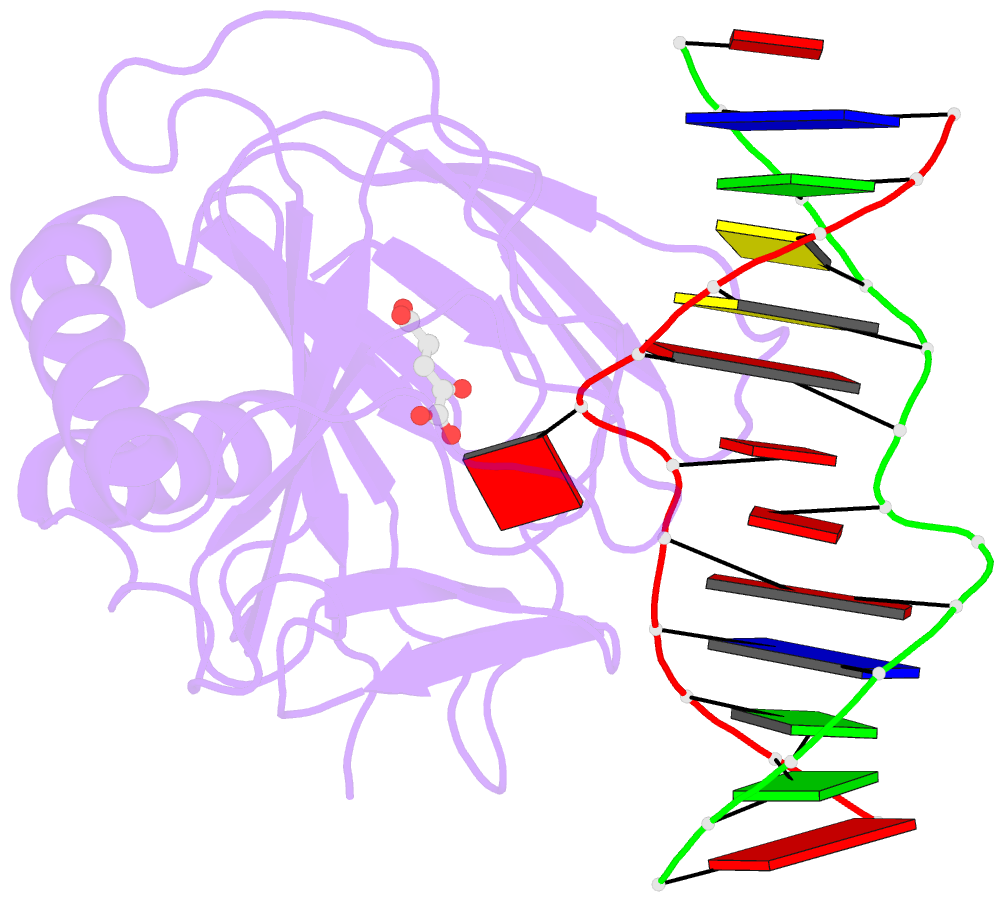
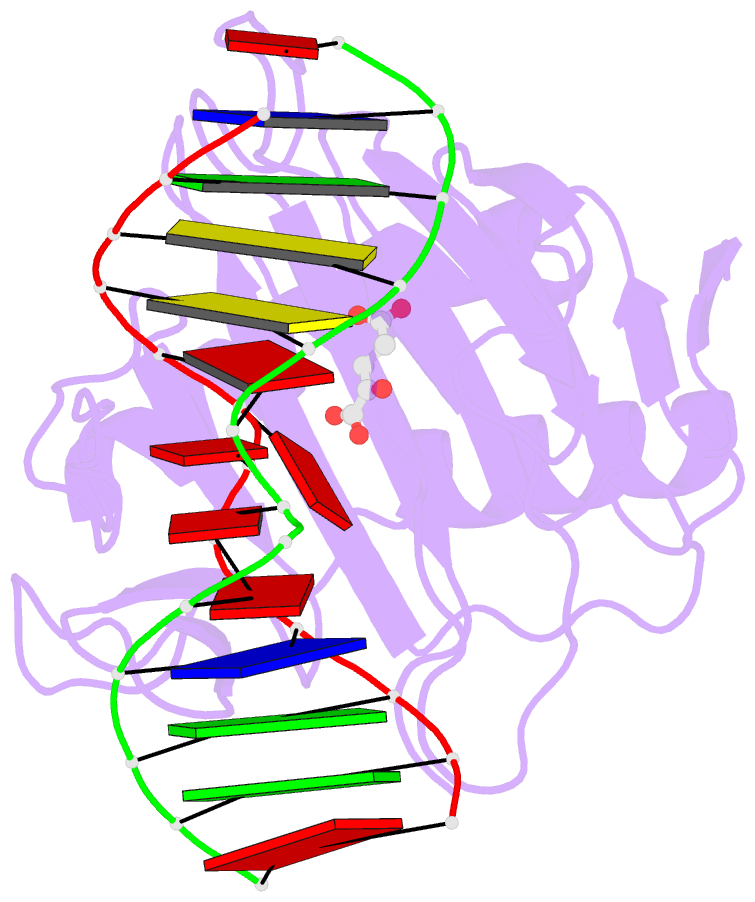
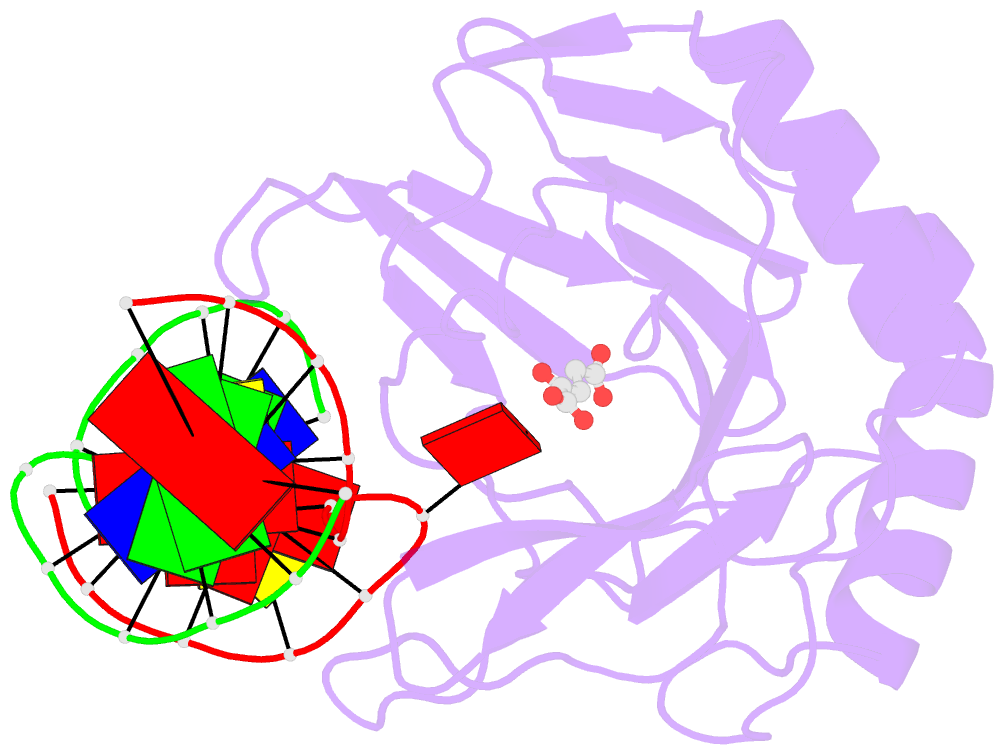
- Crystal structure of alkb protein with cofactors bound to dsDNA containing m6a
- Reference
- Zhu C, Yi C (2014): "Switching Demethylation Activities between AlkB Family RNA/DNA Demethylases through Exchange of Active-Site Residues." Angew.Chem.Int.Ed.Engl., 53, 3659-3662. doi: 10.1002/anie.201310050.
- Abstract
- The AlkB family demethylases AlkB, FTO, and ALKBH5 recognize differentially methylated RNA/DNA substrates, which results in their distinct biological roles. Here we identify key active-site residues that contribute to their substrate specificity. Swapping such active-site residues between the demethylases leads to partially switched demethylation activities. Combined evidence from X-ray structures and enzyme kinetics suggests a role of the active-site residues in substrate recognition. Such a divergent active-site sequence may aid the design of selective inhibitors that can discriminate these homologue RNA/DNA demethylases.