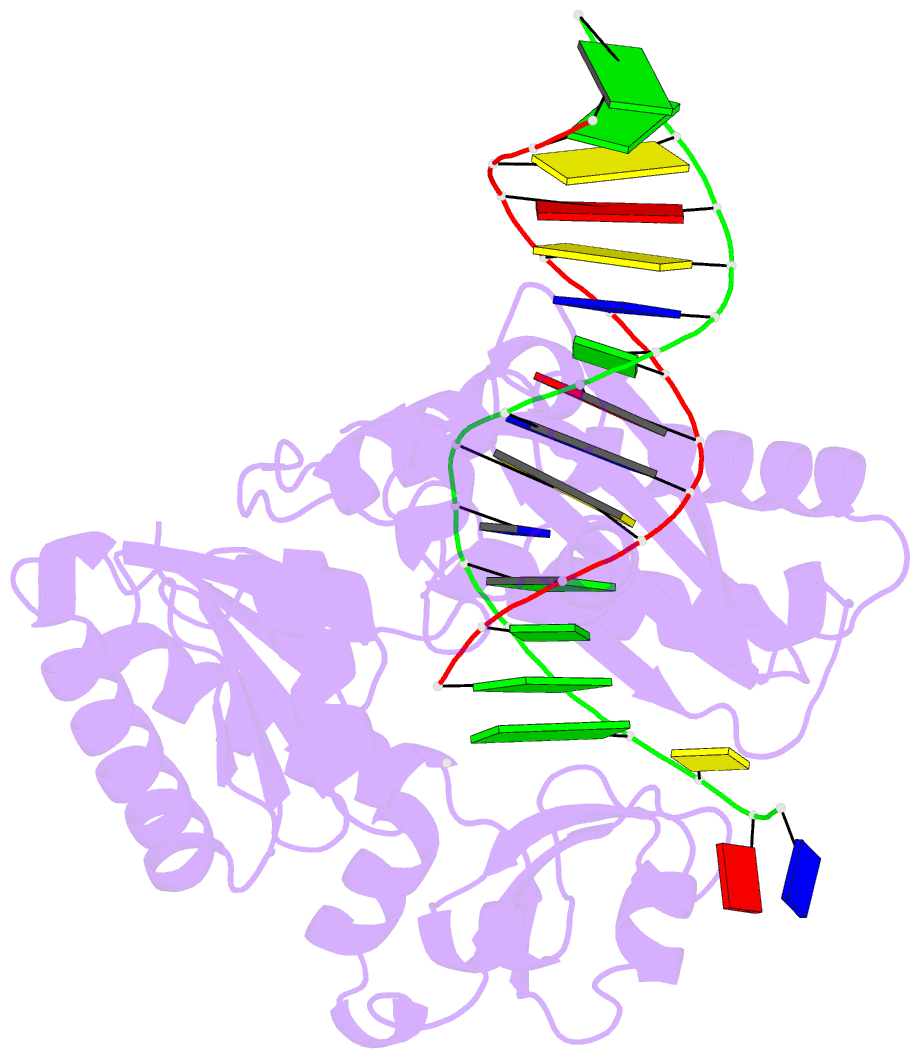
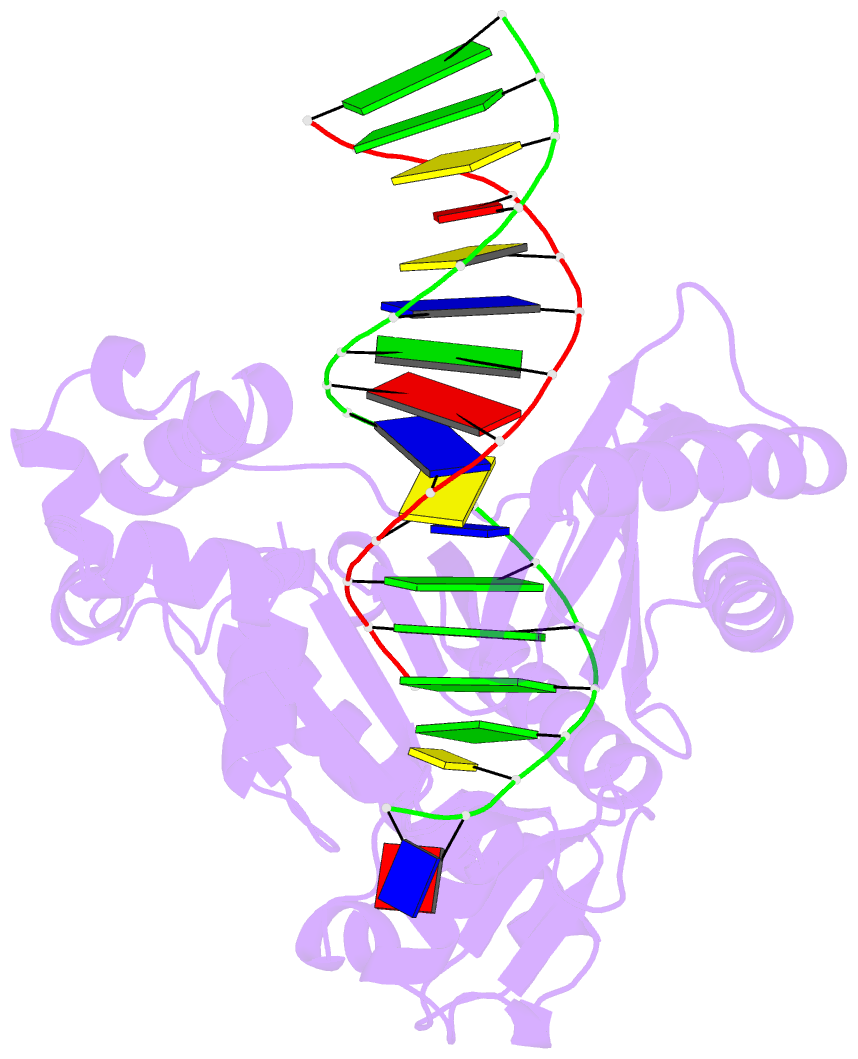
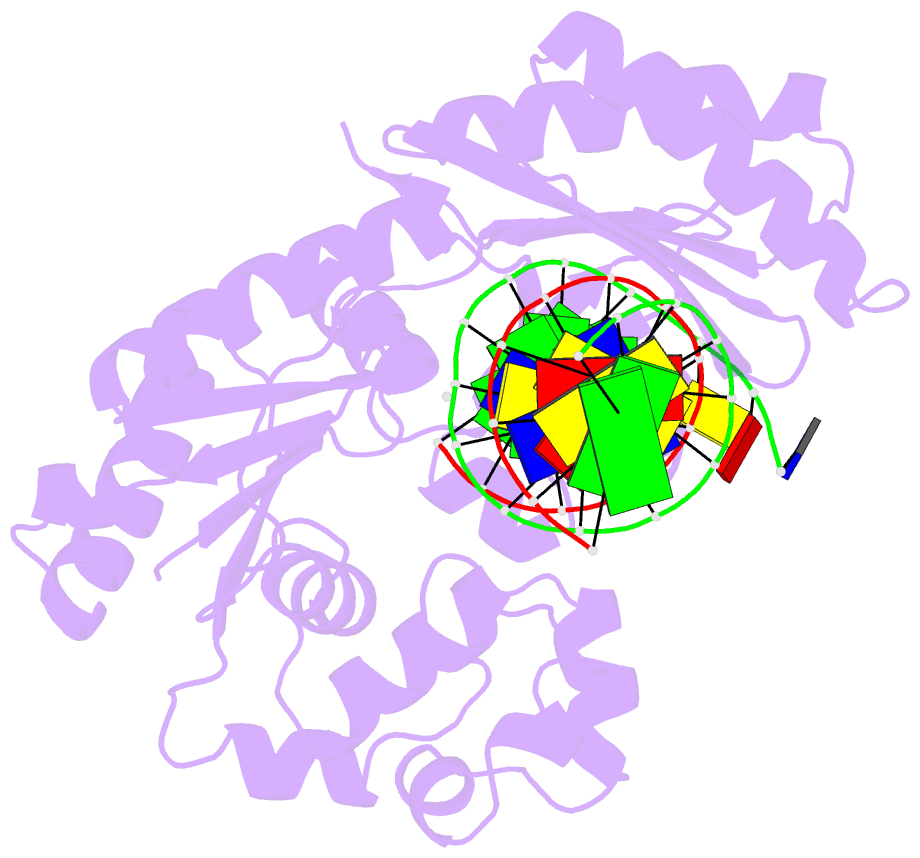
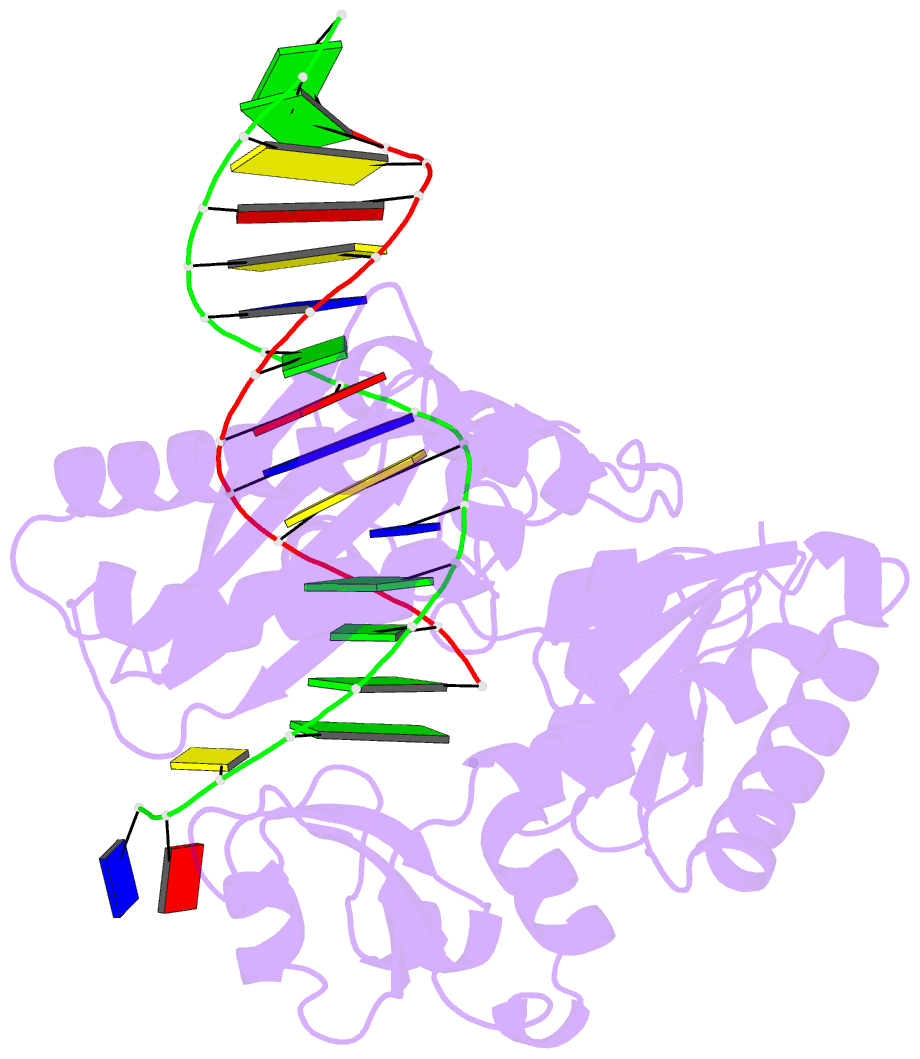
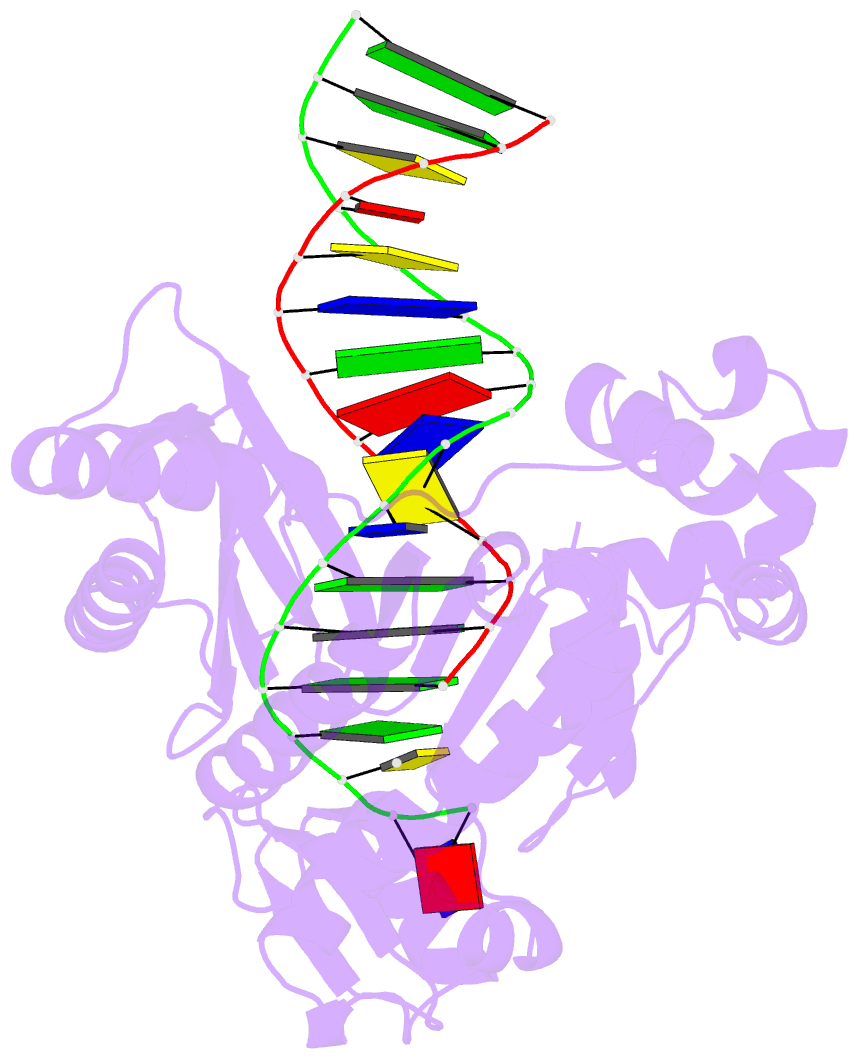
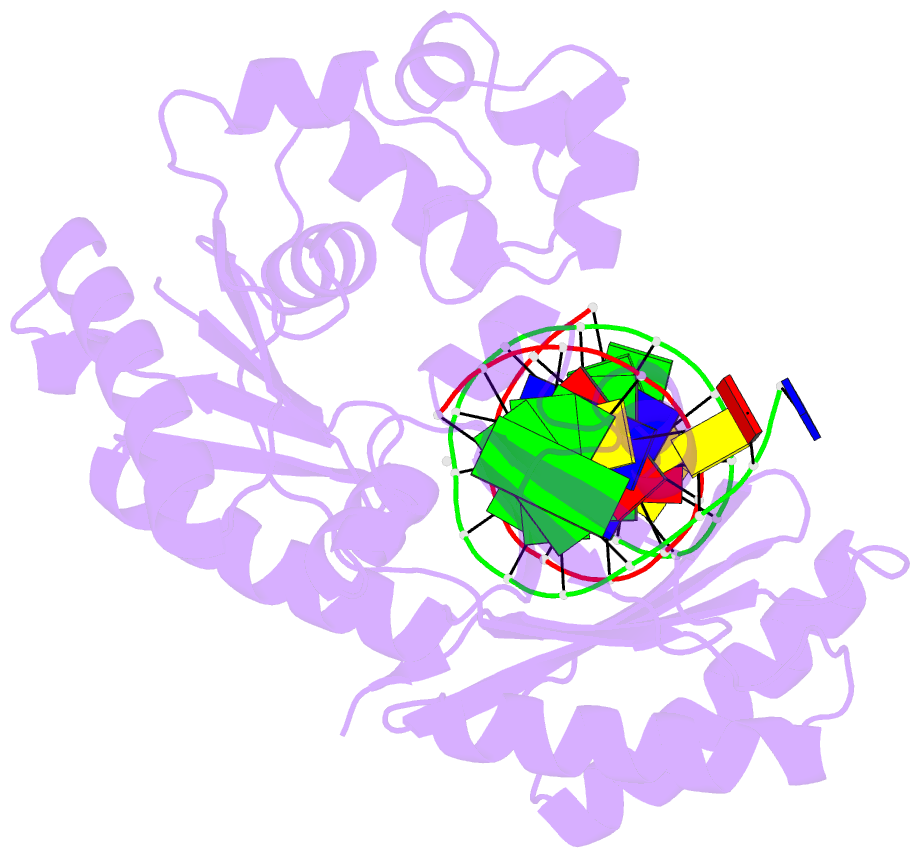
Summary information and primary citation
- PDB-id
- 4nlg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.4 Å)
- Summary
- Y-family DNA polymerase chimera dbh-dpo4(243-245)-dbh
- Reference
- Mukherjee P, Wilson RC, Lahiri I, Pata JD (2014): "Three Residues of the Interdomain Linker Determine the Conformation and Single-base Deletion Fidelity of Y-family Translesion Polymerases." J.Biol.Chem., 289, 6323-6331. doi: 10.1074/jbc.M113.537860.
- Abstract
- Dpo4 and Dbh are from two closely related Sulfolobus species and are well studied archaeal homologues of pol IV, an error prone Y-family polymerase from Escherichia coli. Despite sharing 54% amino acid identity, these polymerases display distinct mutagenic and translesion specificities. Structurally, Dpo4 and Dbh adopt different conformations because of the difference in relative orientation of their N-terminal catalytic and C-terminal DNA binding domains. Using chimeric constructs of these two polymerases, we have previously demonstrated that the interdomain linker is a major determinant of polymerase conformation, base-substitution fidelity, and abasic-site translesion synthesis. Here we find that the interdomain linker also affects the single-base deletion frequency and the mispair extension efficiency of these polymerases. Exchanging just three amino acids in the linkers of Dbh and Dpo4 is sufficient to change the fidelity by up to 30-fold, predominantly by altering the rate of correct (but not incorrect) nucleotide incorporation. Additionally, from a 2.4 Å resolution crystal structure, we have found that the three linker amino acids from Dpo4 are sufficient to allow Dbh to adopt the standard conformation of Dpo4. Thus, a small region of the interdomain linker, located more than 11 Å away from the catalytic residues, determines the fidelity of these Y-family polymerases, by controlling the alignment of substrates at the active site.