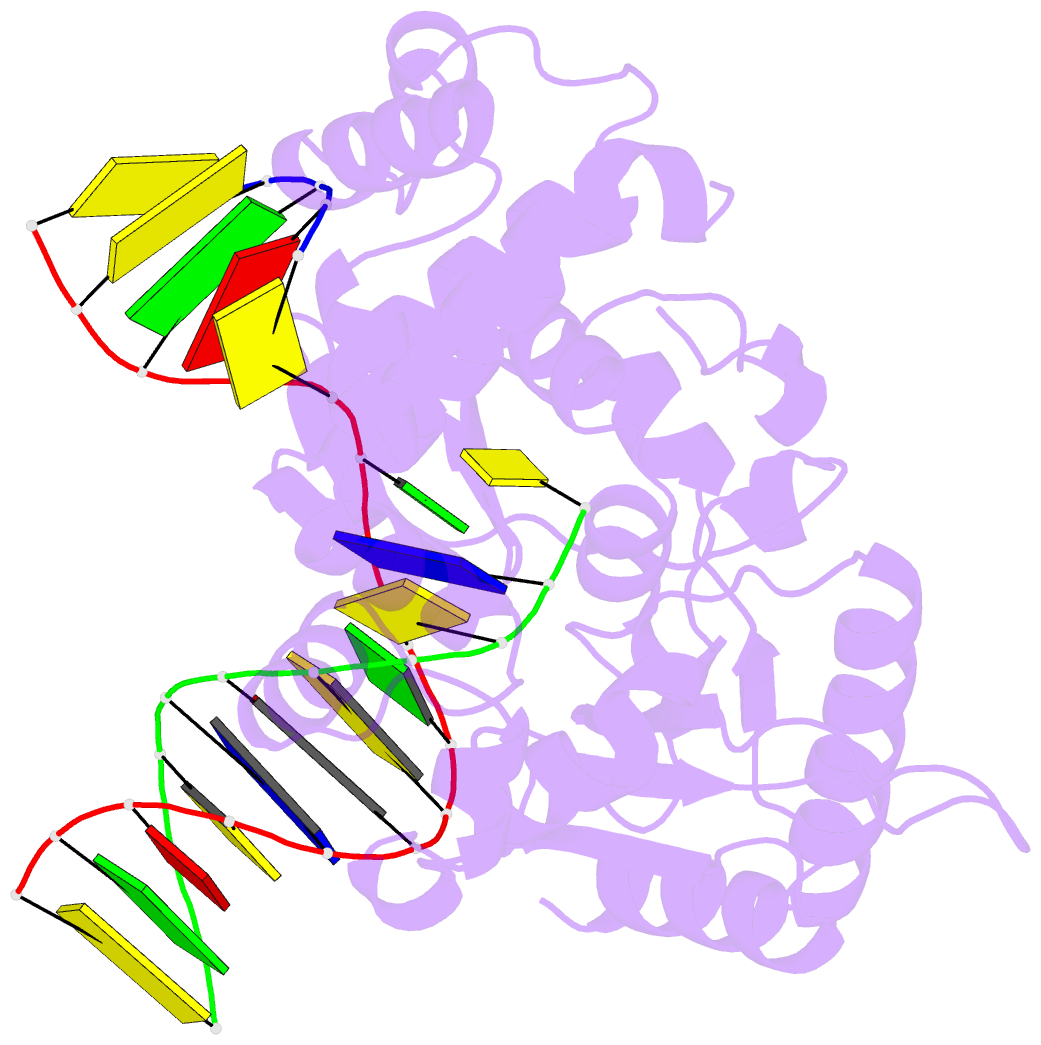
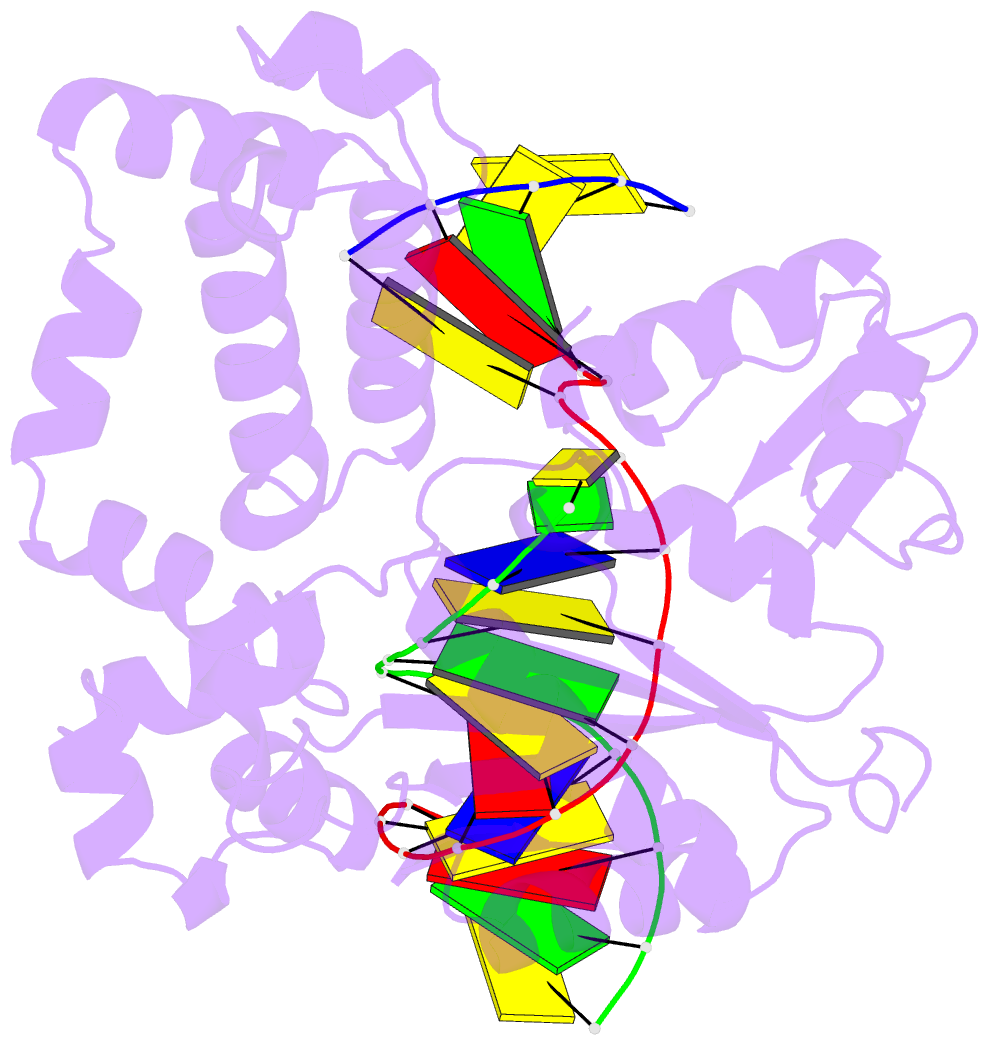
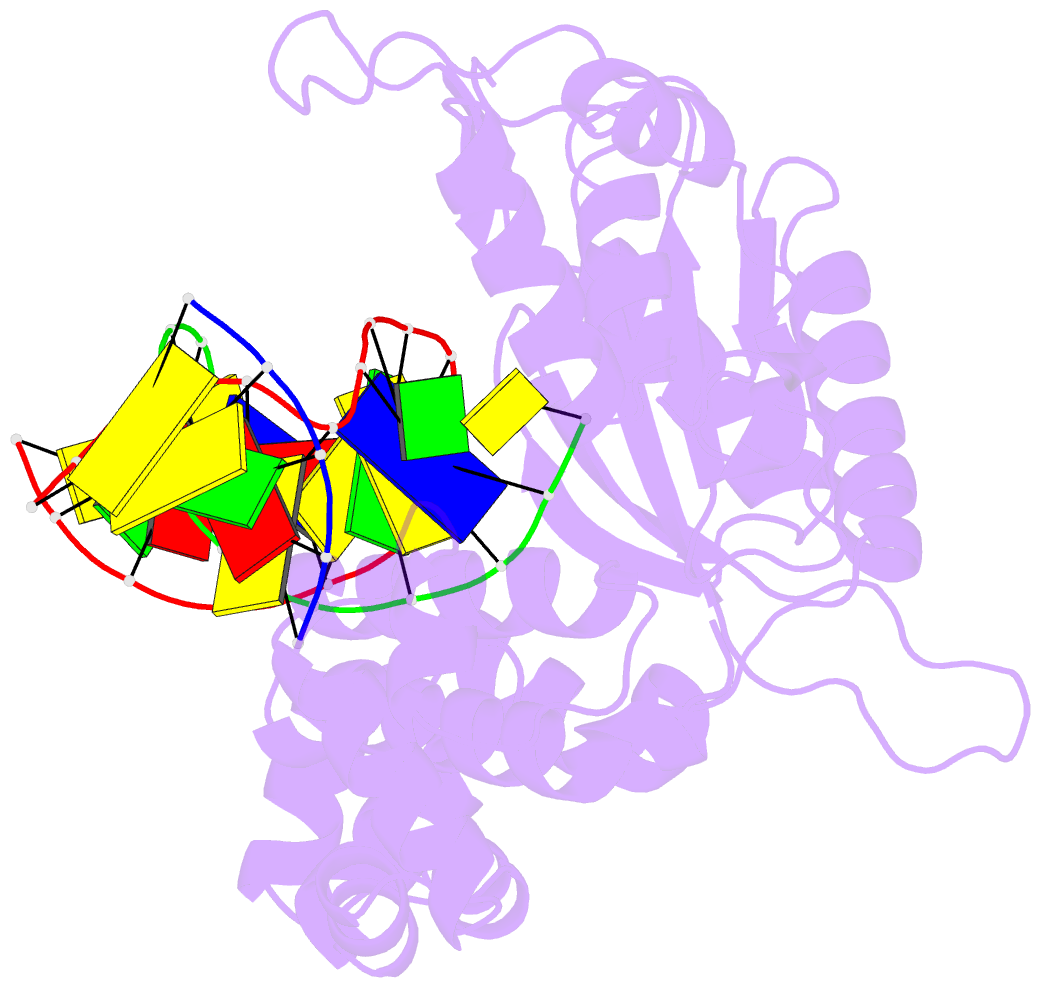
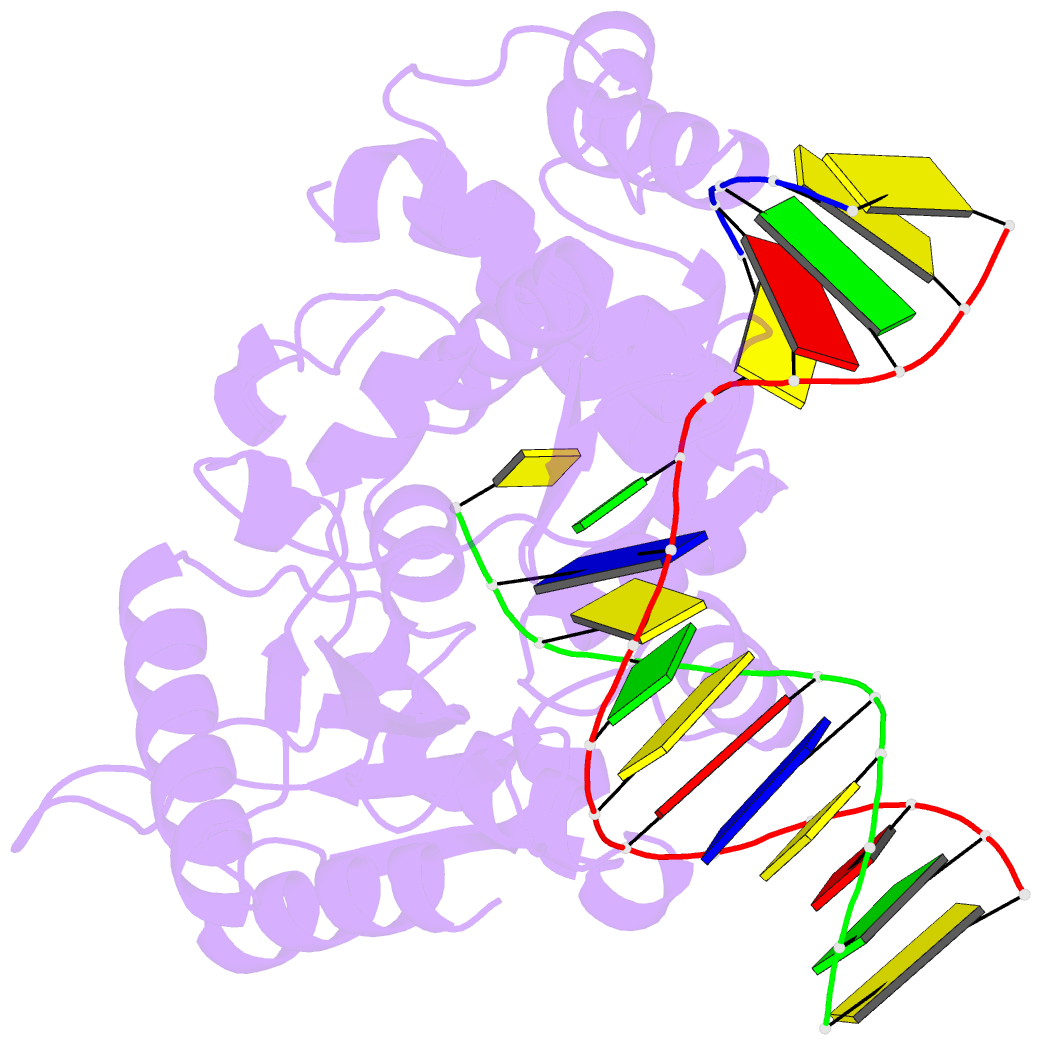
Summary information and primary citation
- PDB-id
- 4nln; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase, lyase-DNA
- Method
- X-ray (2.261 Å)
- Summary
- Structure of human DNA polymerase beta complexed with nicked DNA containing a template 8brg and incoming ctp
- Reference
- Koag MC, Min K, Lee S (2014): "Structural basis for promutagenicity of 8-halogenated Guanine." J.Biol.Chem., 289, 6289-6298. doi: 10.1074/jbc.M113.537803.
- Abstract
- 8-Halogenated guanine (haloG), a major DNA adduct formed by reactive halogen species during inflammation, is a promutagenic lesion that promotes misincorporation of G opposite the lesion by various DNA polymerases. Currently, the structural basis for such misincorporation is unknown. To gain insights into the mechanism of misincorporation across haloG by polymerase, we determined seven x-ray structures of human DNA polymerase β (polβ) bound to DNA bearing 8-bromoguanine (BrG). We determined two pre-catalytic ternary complex structures of polβ with an incoming nonhydrolyzable dGTP or dCTP analog paired with templating BrG. We also determined five binary complex structures of polβ in complex with DNA containing BrG·C/T at post-insertion and post-extension sites. In the BrG·dGTP ternary structure, BrG adopts syn conformation and forms Hoogsteen base pairing with the incoming dGTP analog. In the BrG·dCTP ternary structure, BrG adopts anti conformation and forms Watson-Crick base pairing with the incoming dCTP analog. In addition, our polβ binary post-extension structures show Hoogsteen BrG·G base pair and Watson-Crick BrG·C base pair. Taken together, the first structures of haloG-containing DNA bound to a protein indicate that both BrG·G and BrG·C base pairs are accommodated in the active site of polβ. Our structures suggest that Hoogsteen-type base pairing between G and C8-modified G could be accommodated in the active site of a DNA polymerase, promoting G to C mutation.