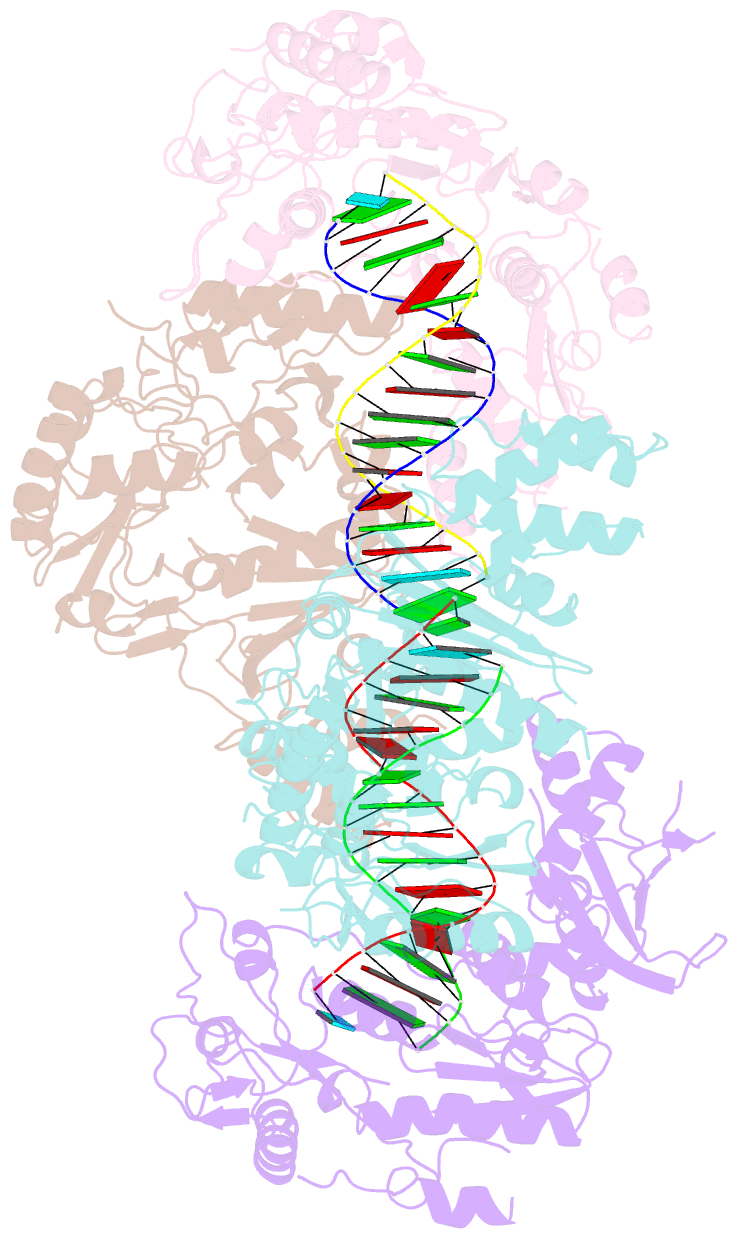
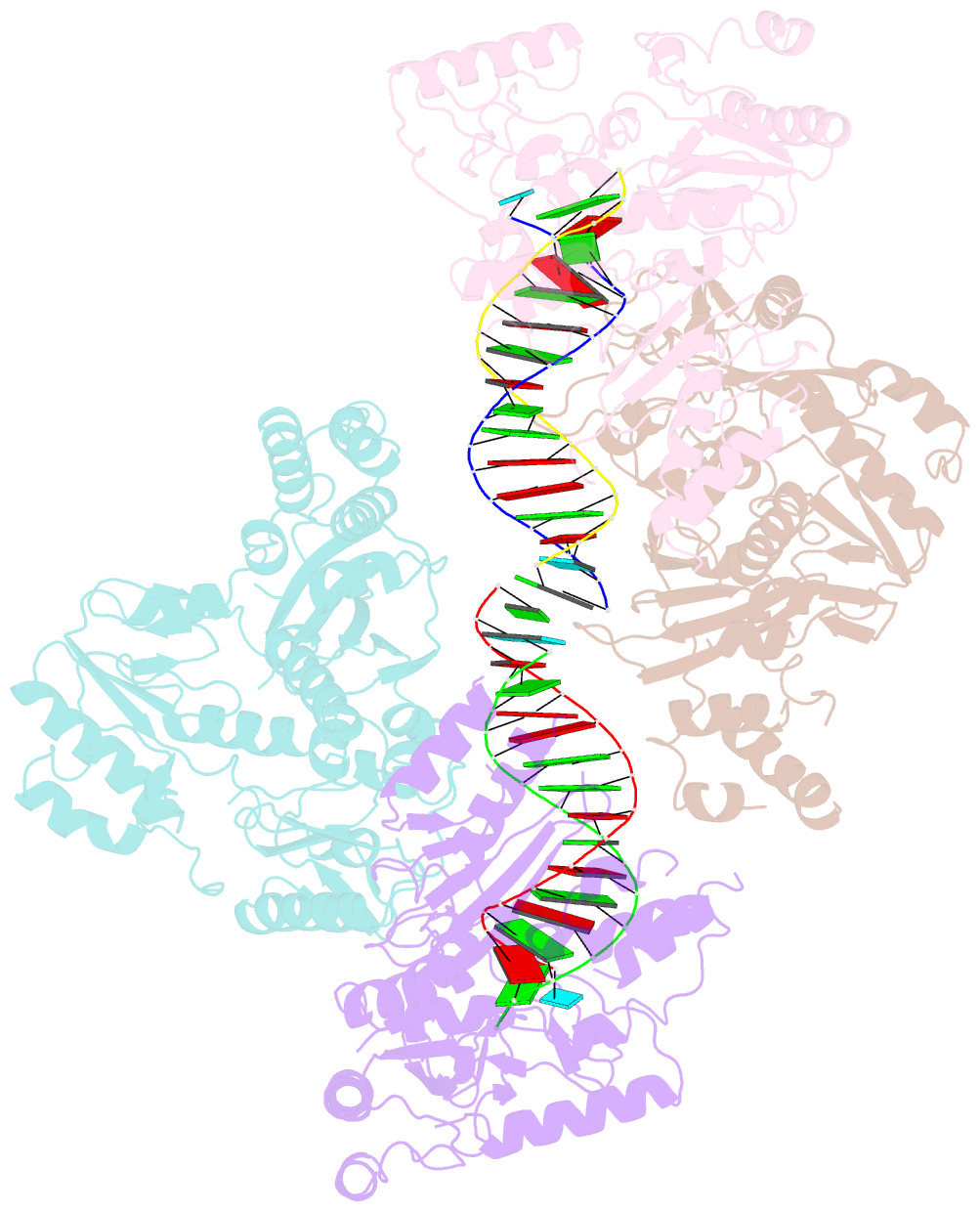
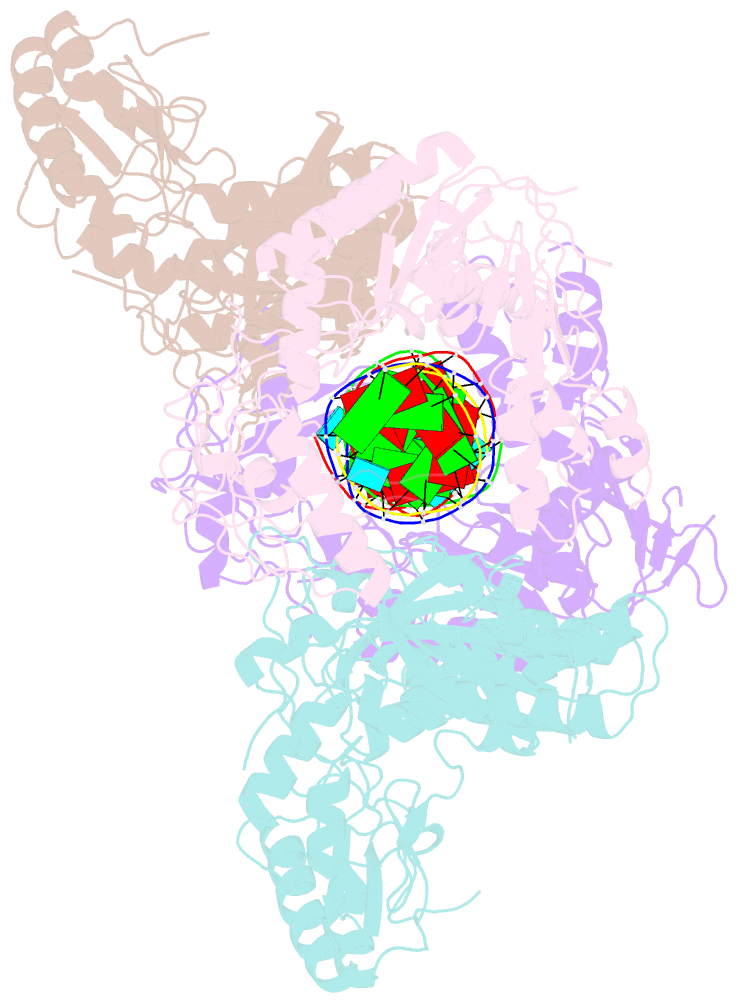
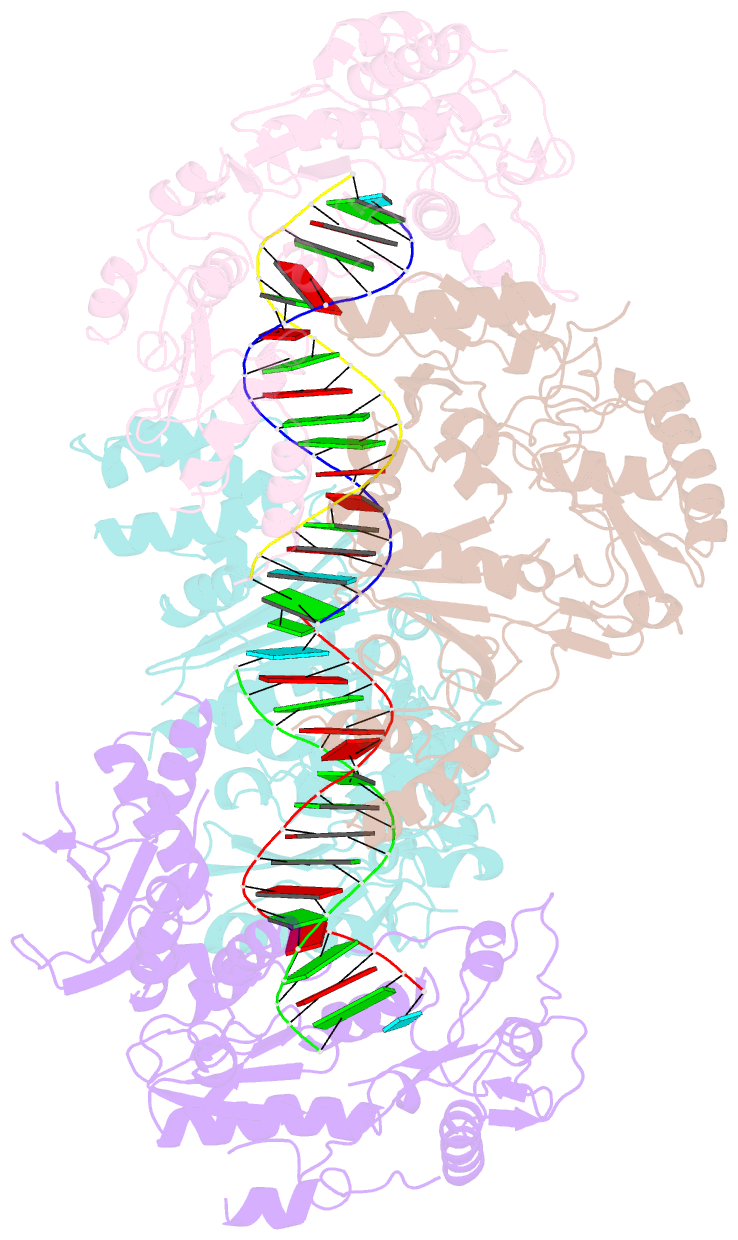
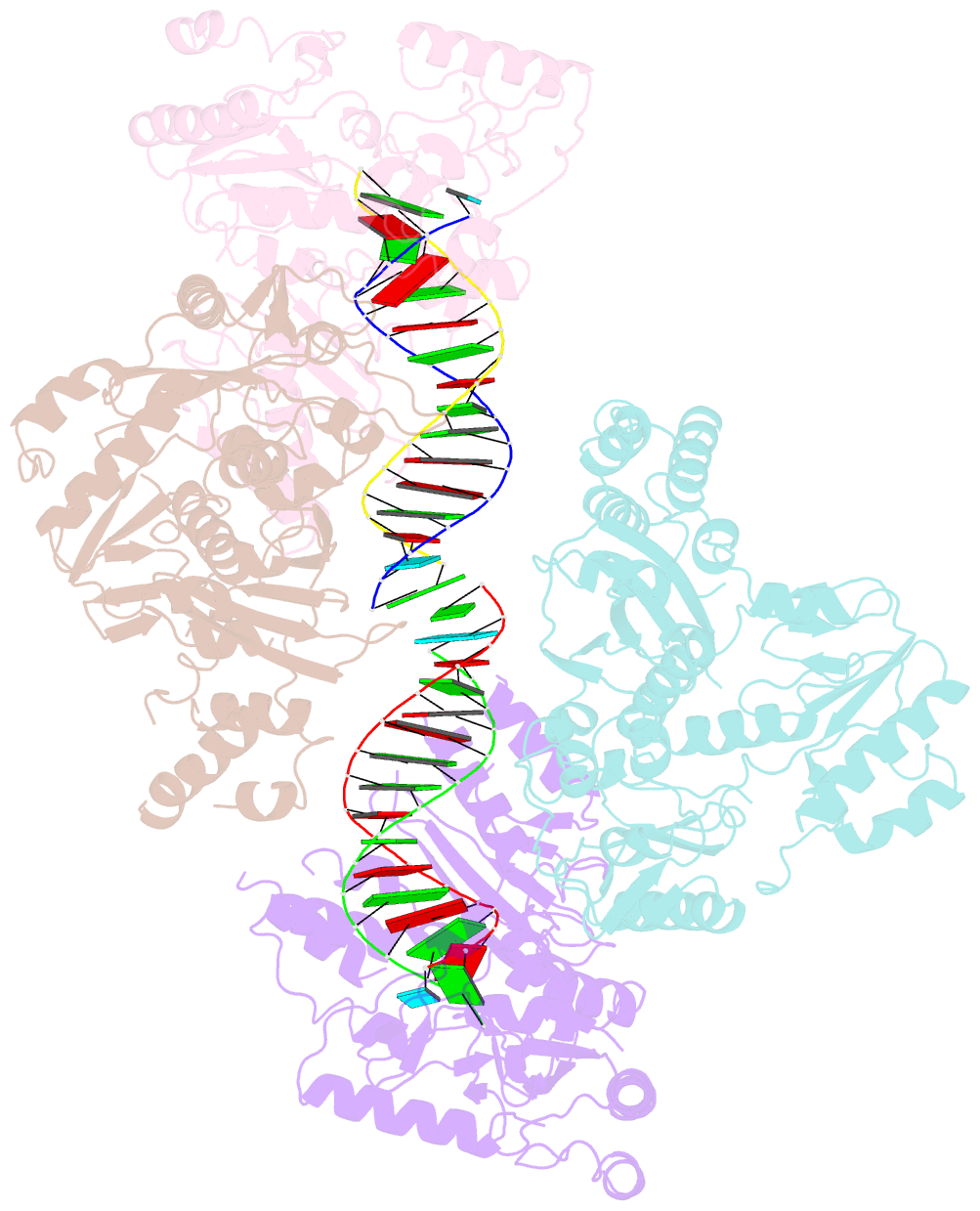
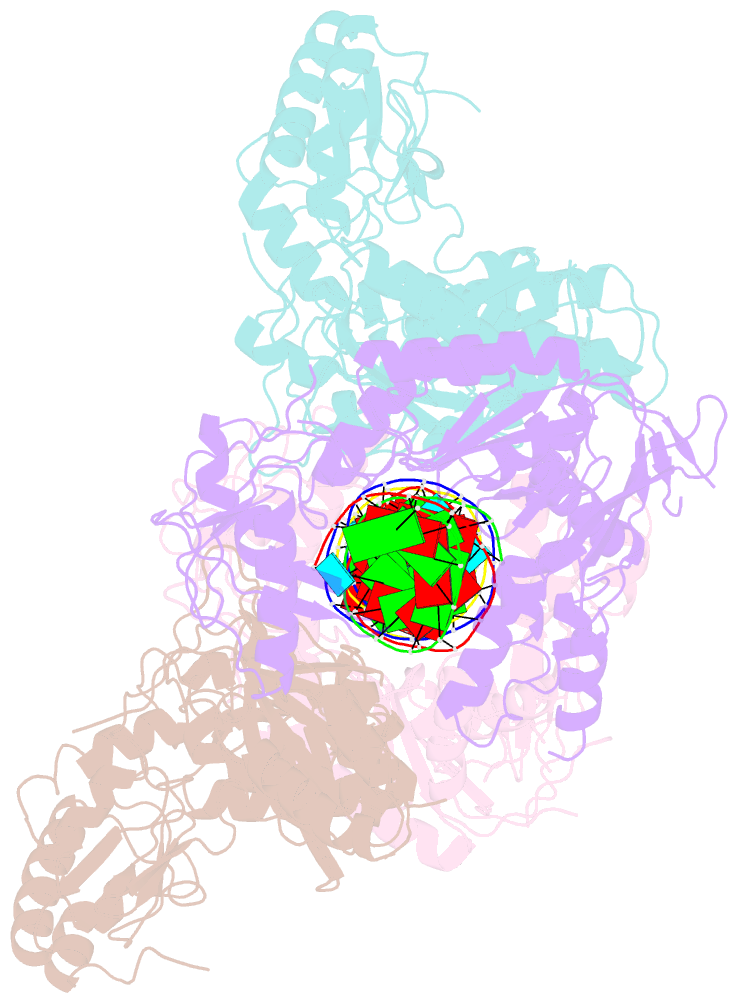
Summary information and primary citation
- PDB-id
- 4ol8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase, hydrolase-RNA-DNA
- Method
- X-ray (3.1 Å)
- Summary
- Ty3 reverse transcriptase bound to DNA-RNA
- Reference
- Nowak E, Miller JT, Bona MK, Studnicka J, Szczepanowski RH, Jurkowski J, Le Grice SF, Nowotny M (2014): "Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry." Nat.Struct.Mol.Biol., 21, 389-396. doi: 10.1038/nsmb.2785.
- Abstract
- Retrotransposons are a class of mobile genetic elements that replicate by converting their single-stranded RNA intermediate to double-stranded DNA through the combined DNA polymerase and ribonuclease H (RNase H) activities of the element-encoded reverse transcriptase (RT). Although a wealth of structural information is available for lentiviral and gammaretroviral RTs, equivalent studies on counterpart enzymes of long terminal repeat (LTR)-containing retrotransposons, from which they are evolutionarily derived, is lacking. In this study, we report the first crystal structure of a complex of RT from the Saccharomyces cerevisiae LTR retrotransposon Ty3 in the presence of its polypurine tract-containing RNA-DNA hybrid. In contrast to its retroviral counterparts, Ty3 RT adopts an asymmetric homodimeric architecture whose assembly is substrate dependent. Moreover, our structure and biochemical data suggest that the RNase H and DNA polymerase activities are contributed by individual subunits of the homodimer.