Summary information and primary citation
- PDB-id
- 4oog; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.5 Å)
- Summary
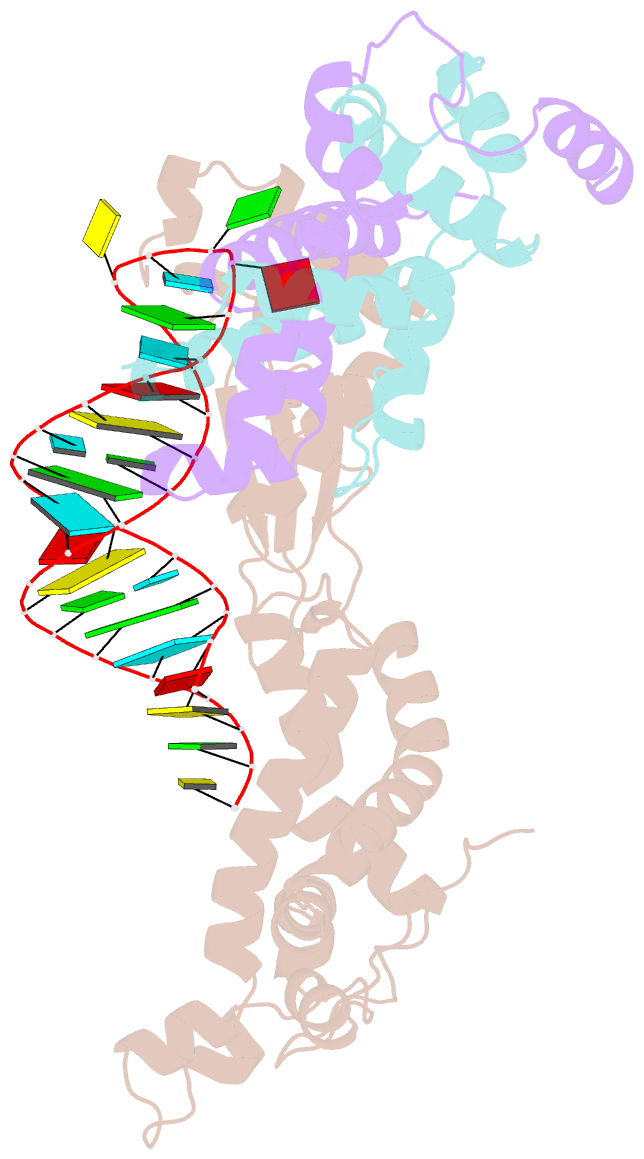
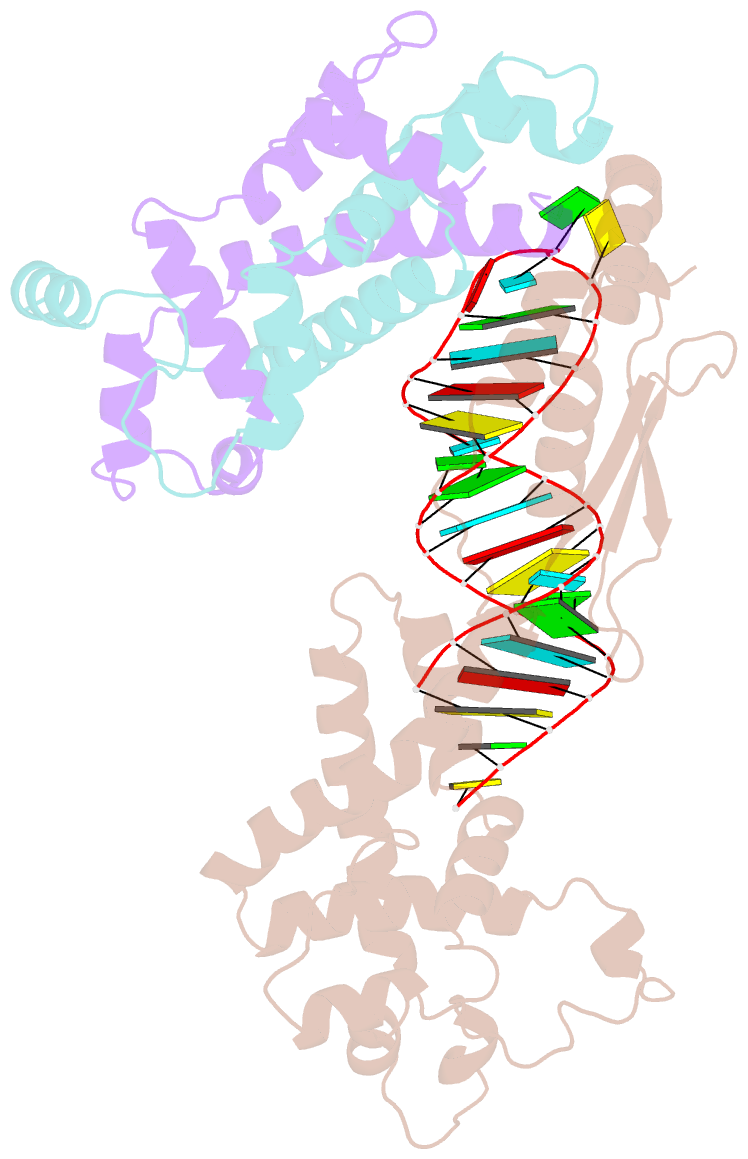
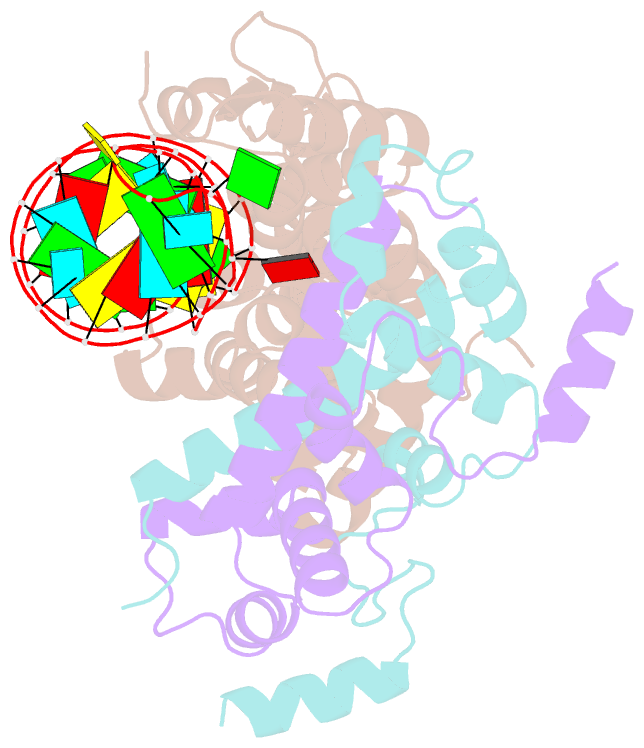
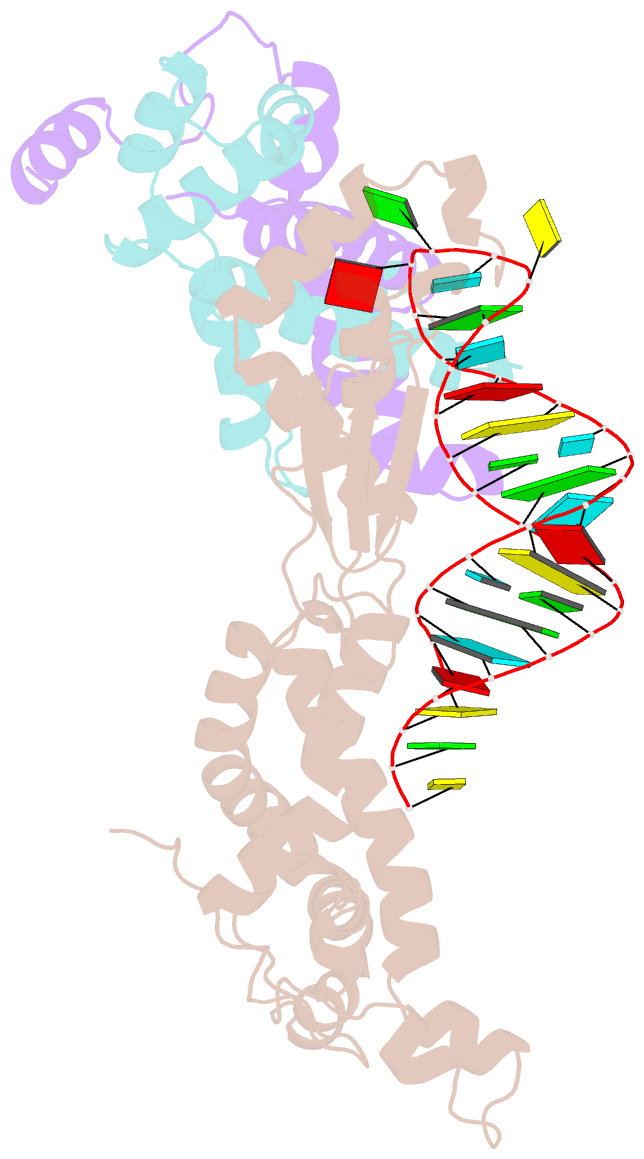
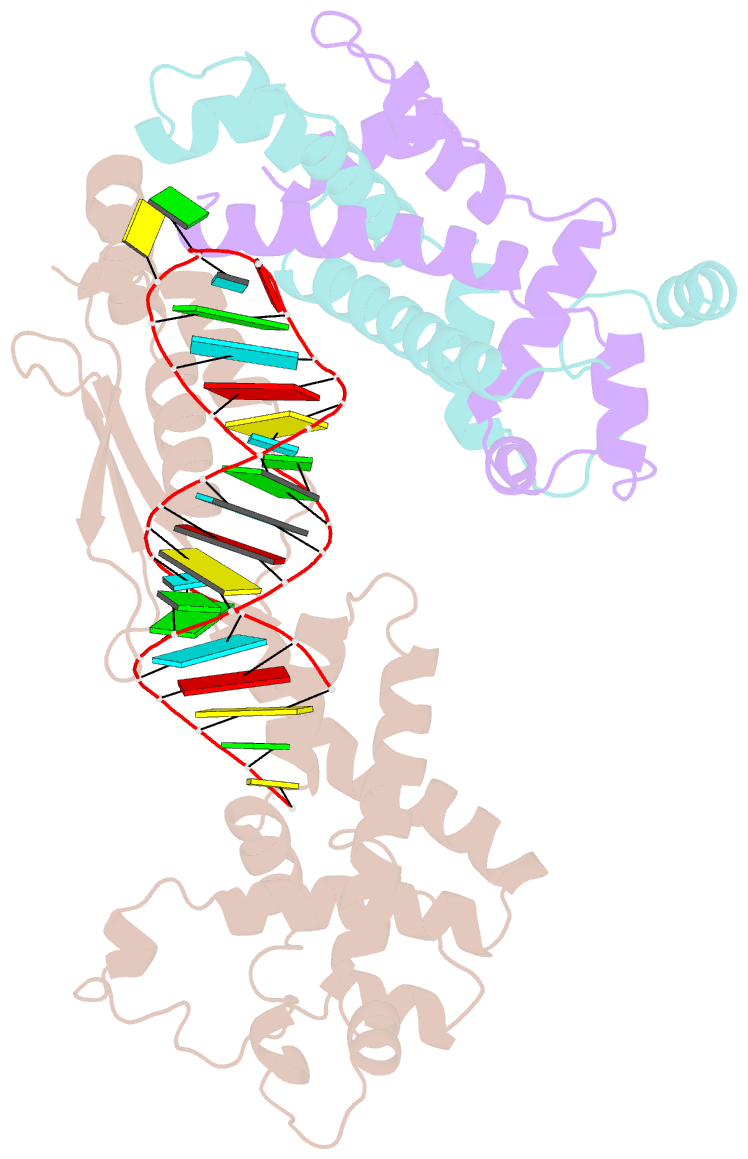
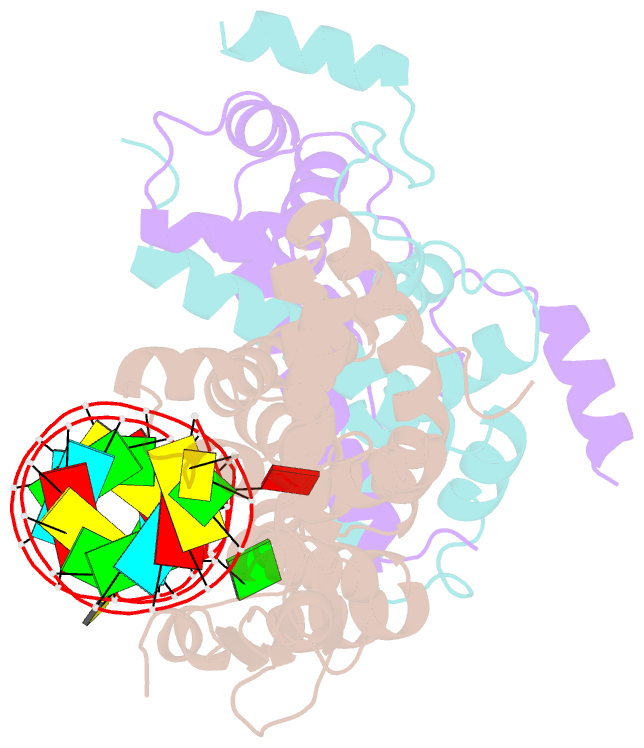
- Crystal structure of yeast rnase iii (rnt1p) complexed with the product of dsrna processing
- Reference
- Liang YH, Lavoie M, Comeau MA, Abou Elela S, Ji X (2014): "Structure of a Eukaryotic RNase III Postcleavage Complex Reveals a Double-Ruler Mechanism for Substrate Selection." Mol.Cell, 54, 431-444. doi: 10.1016/j.molcel.2014.03.006.
- Abstract
- Ribonuclease III (RNase III) enzymes are a family of double-stranded RNA (dsRNA)-specific endoribonucleases required for RNA maturation and gene regulation. Prokaryotic RNase III enzymes have been well characterized, but how eukaryotic RNase IIIs work is less clear. Here, we describe the structure of the Saccharomyces cerevisiae RNase III (Rnt1p) postcleavage complex and explain why Rnt1p binds to RNA stems capped with an NGNN tetraloop. The structure shows specific interactions between a structural motif located at the end of the Rnt1p dsRNA-binding domain (dsRBD) and the guanine nucleotide in the second position of the loop. Strikingly, structural and biochemical analyses indicate that the dsRBD and N-terminal domains (NTDs) of Rnt1p function as two rulers that measure the distance between the tetraloop and the cleavage site. These findings provide a framework for understanding eukaryotic RNase IIIs.