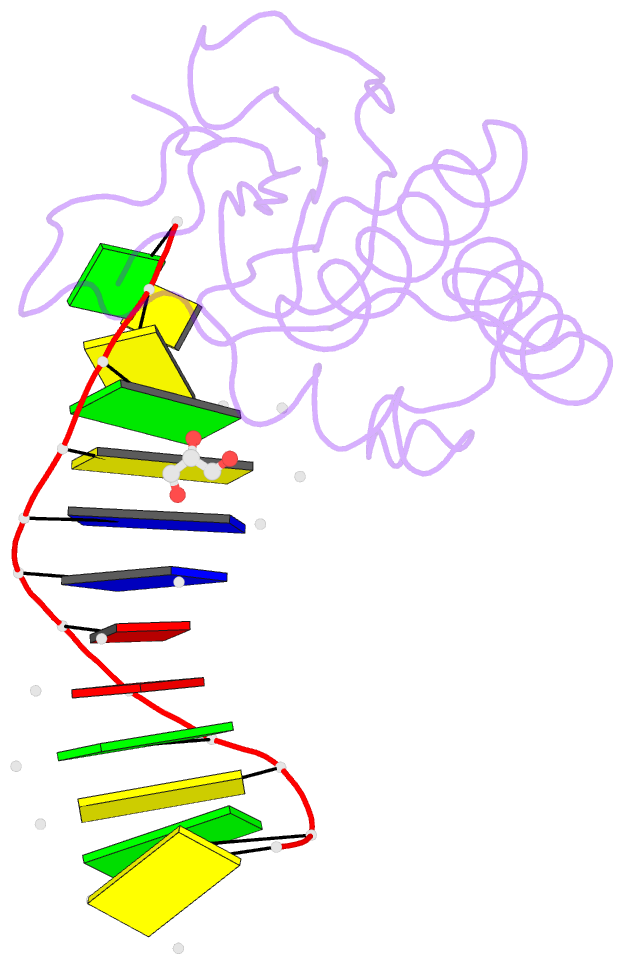
Summary information and primary citation
- PDB-id
- 4opj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.541 Å)
- Summary
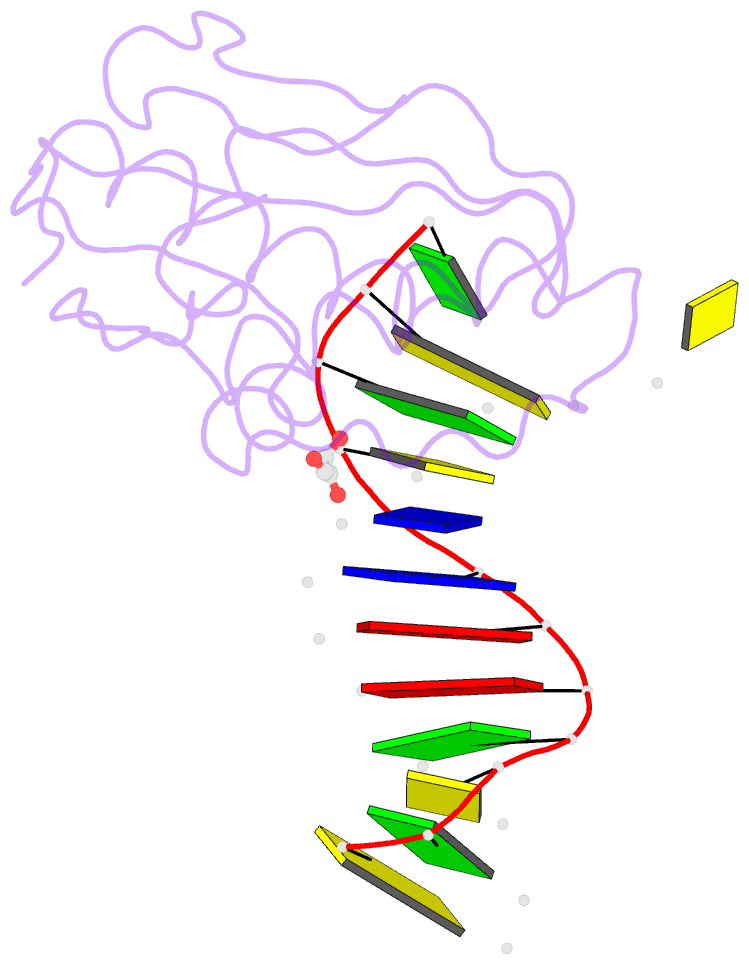
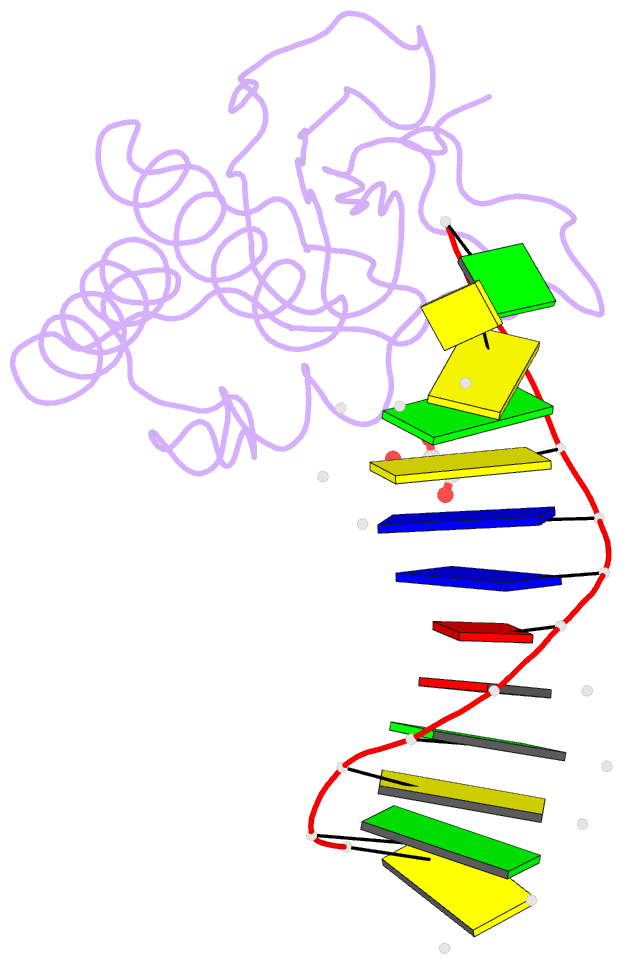
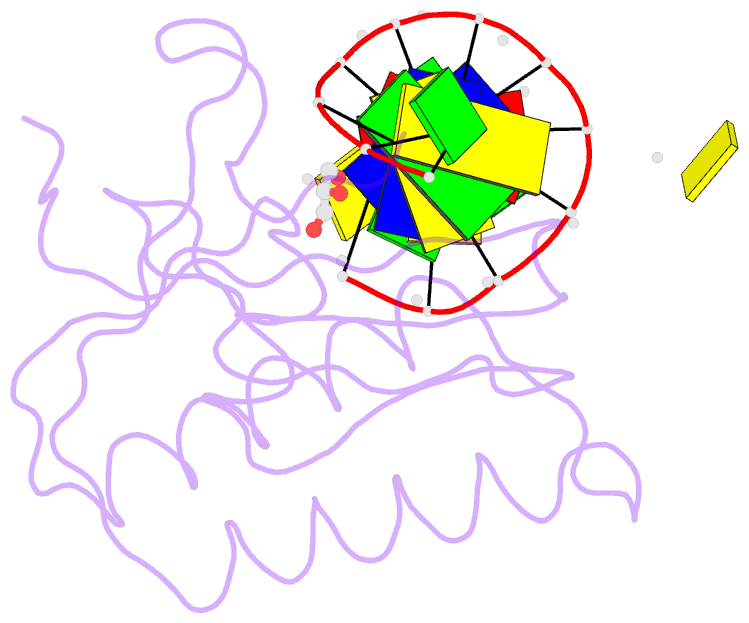
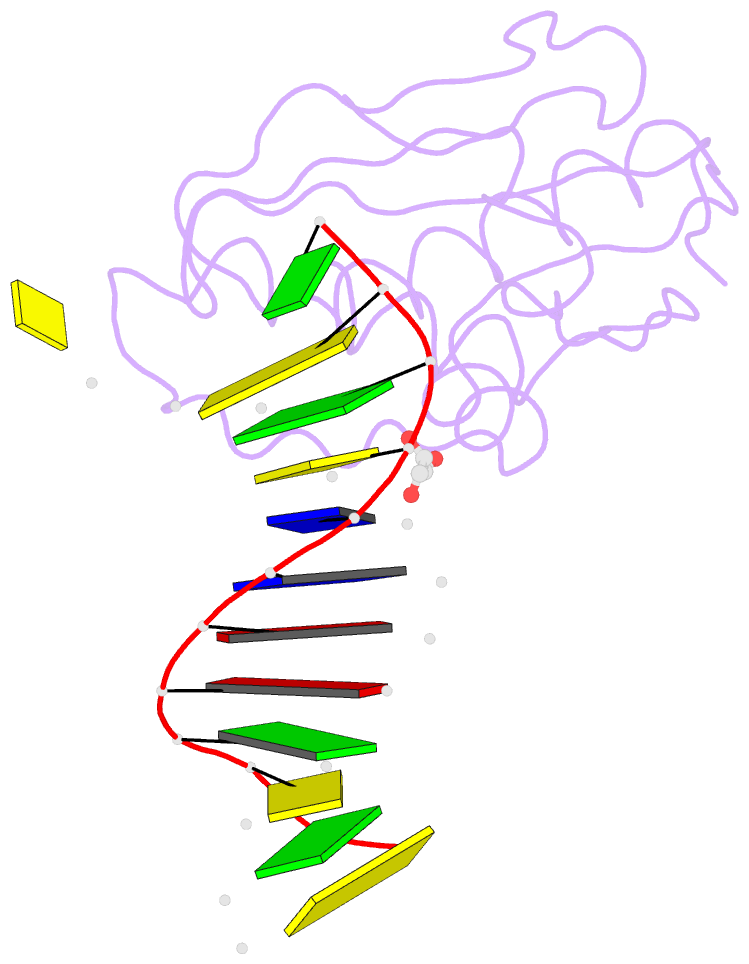
- Bh-rnaseh:tcda-DNA complex
- Reference
- Egli M, Pallan PS: "Generating Crystallographic Models of DNA Dodecamers from Structures of RNase H:DNA Complexes." Methods Mol.Biol., 1320, 111-126. doi: 10.1007/978-1-4939-2763-0_8.
- Abstract
- The DNA dodecamer 5'-d(CGCGAATTCGCG)-3' is arguably the best studied oligonucleotide and crystal structures of duplexes with this sequence account for a considerable portion of the total number of oligo-2'-deoxynucleotide structures determined over the last 30 years. The dodecamer has commonly served as a template to analyze the effects of sequence on DNA conformation, the conformational properties of chemically modified nucleotides, DNA-ligand interactions as well as water structure and DNA-cation binding. Although molecular replacement is the phasing method of choice given the large number of available models of the dodecamer, this strategy often fails as a result of conformational changes caused by chemical modification, mismatch pairs, or differing packing modes. Here, we describe an alternative approach to determine crystal structures of the dodecamer in cases where molecular replacement does not produce a solution or when crystals of the DNA alone cannot be grown. It is based on the discovery that many dodecamers of the above sequence can be readily co-crystallized with Bacillus halodurans RNase H, whereby the enzyme is unable to cleave the DNA. Determination of the structure of the complex using the protein portion as the search model yields a structural model of the DNA. Provided crystals of the DNA alone are also available, the DNA model from the complex then enables phasing their structures by molecular replacement.