Summary information and primary citation
- PDB-id
- 4ou7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.83 Å)
- Summary
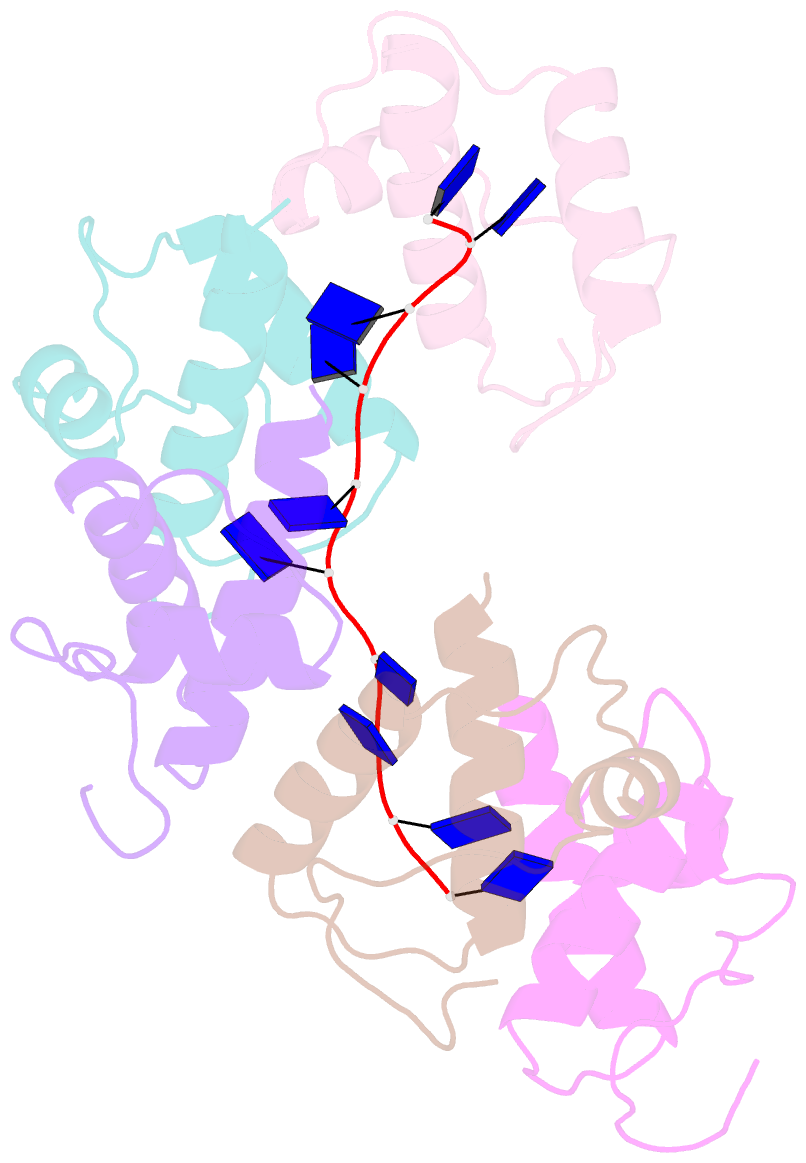
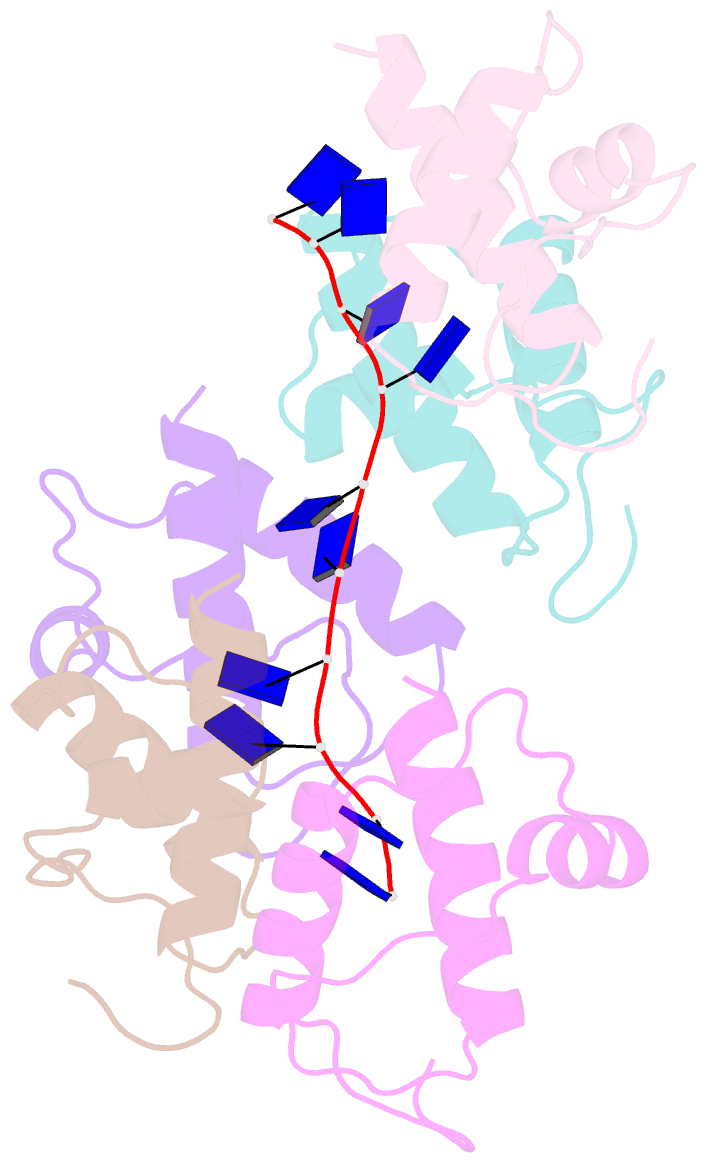
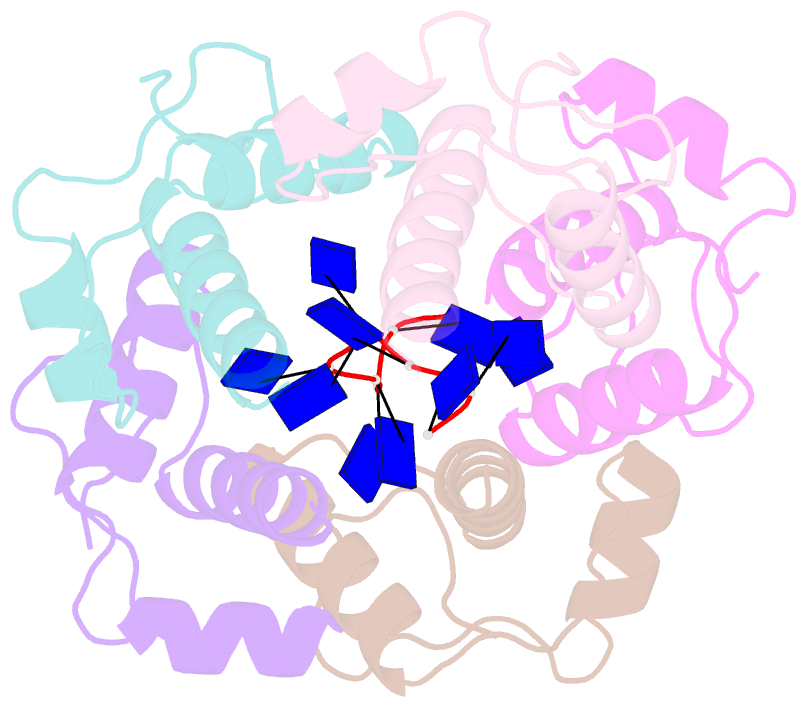
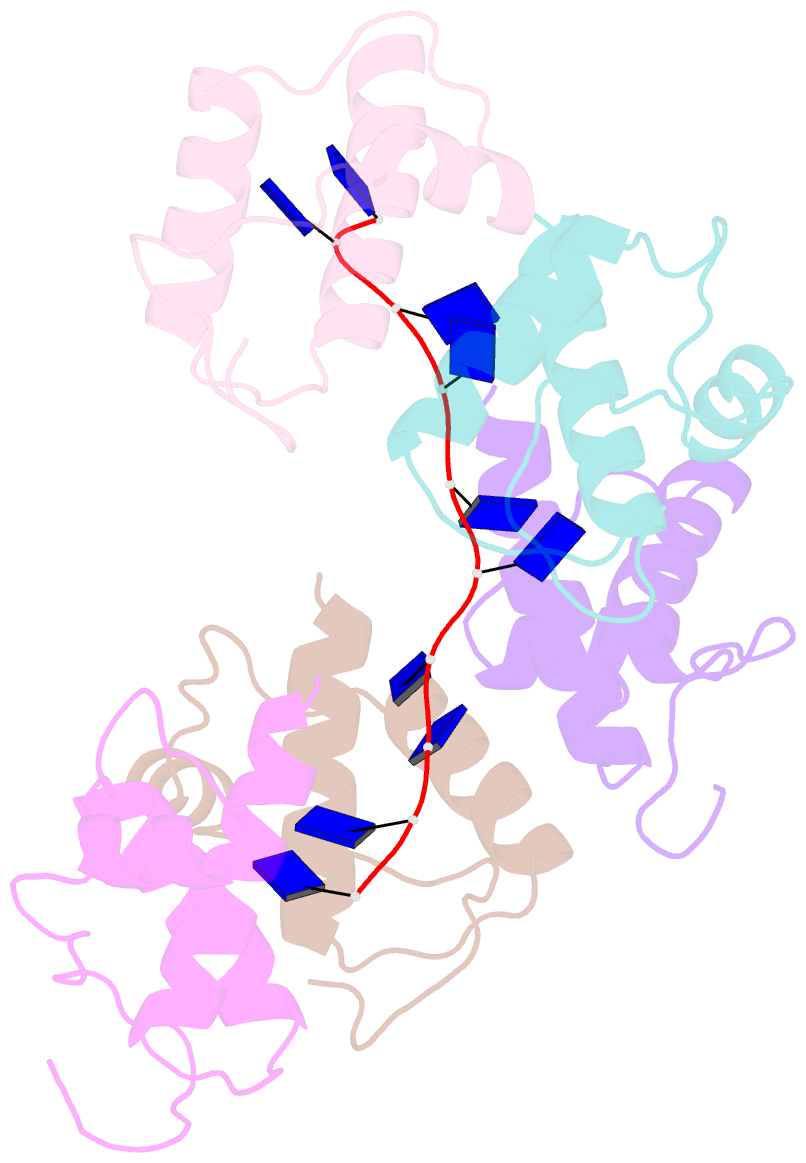
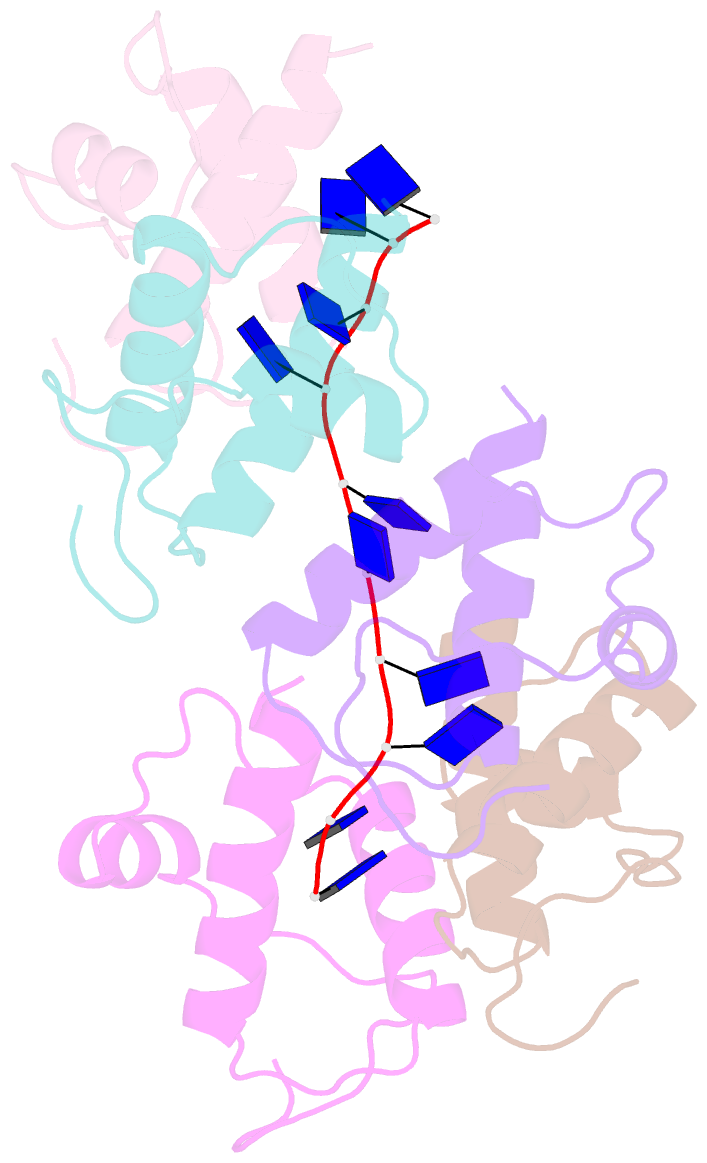
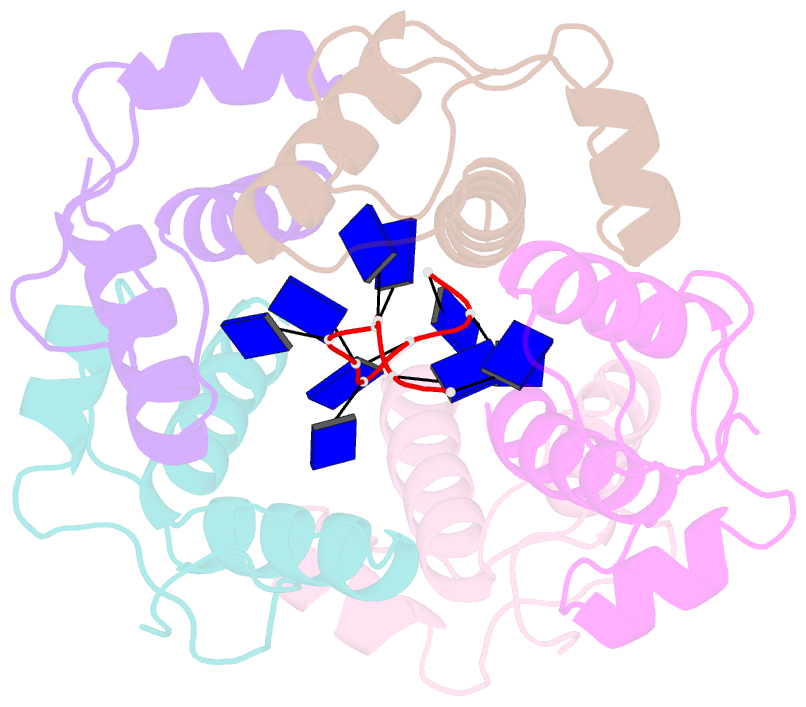
- Crystal structure of dnat84-153-dt10 ssDNA complex reveals a novel single-stranded DNA binding mode
- Reference
- Liu Z, Chen P, Wang X, Cai G, Niu L, Teng M, Li X (2014): "Crystal structure of DnaT84-153-dT10 ssDNA complex reveals a novel single-stranded DNA binding mode." Nucleic Acids Res., 42, 9470-9483. doi: 10.1093/nar/gku633.
- Abstract
- DnaT is a primosomal protein that is required for the stalled replication fork restart in Escherichia coli. As an adapter, DnaT mediates the PriA-PriB-ssDNA ternary complex and the DnaB/C complex. However, the fundamental function of DnaT during PriA-dependent primosome assembly is still a black box. Here, we report the 2.83 Å DnaT(84-153)-dT10 ssDNA complex structure, which reveals a novel three-helix bundle single-stranded DNA binding mode. Based on binding assays and negative-staining electron microscopy results, we found that DnaT can bind to phiX 174 ssDNA to form nucleoprotein filaments for the first time, which indicates that DnaT might function as a scaffold protein during the PriA-dependent primosome assembly. In combination with biochemical analysis, we propose a cooperative mechanism for the binding of DnaT to ssDNA and a possible model for the assembly of PriA-PriB-ssDNA-DnaT complex that sheds light on the function of DnaT during the primosome assembly and stalled replication fork restart. This report presents the first structure of the DnaT C-terminal complex with ssDNA and a novel model that explains the interactions between the three-helix bundle and ssDNA.