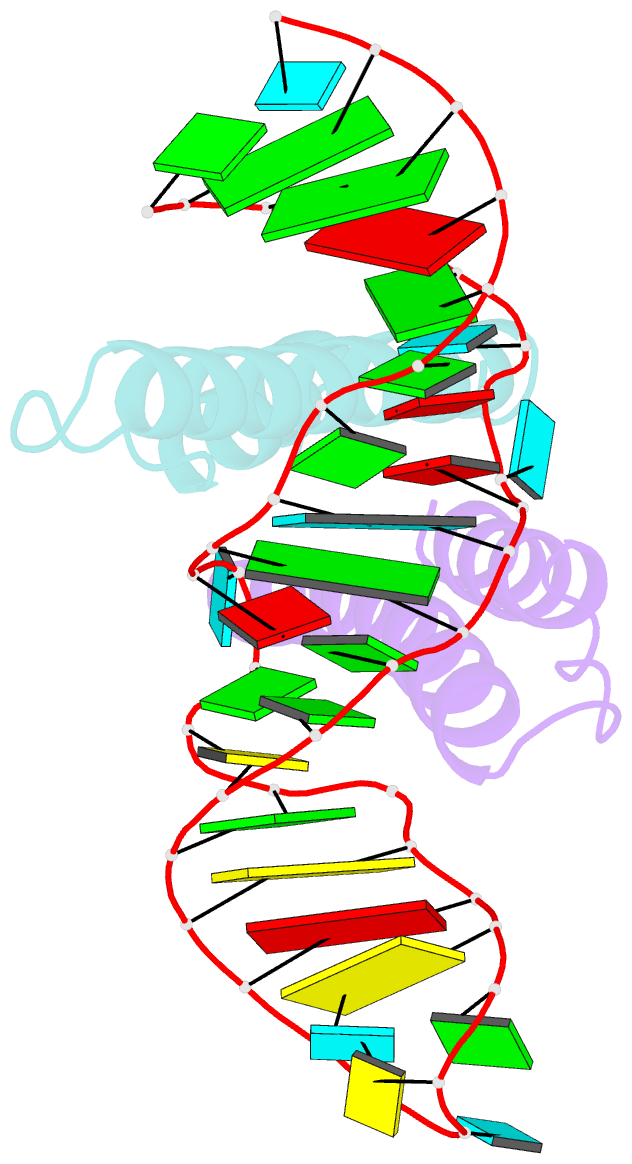
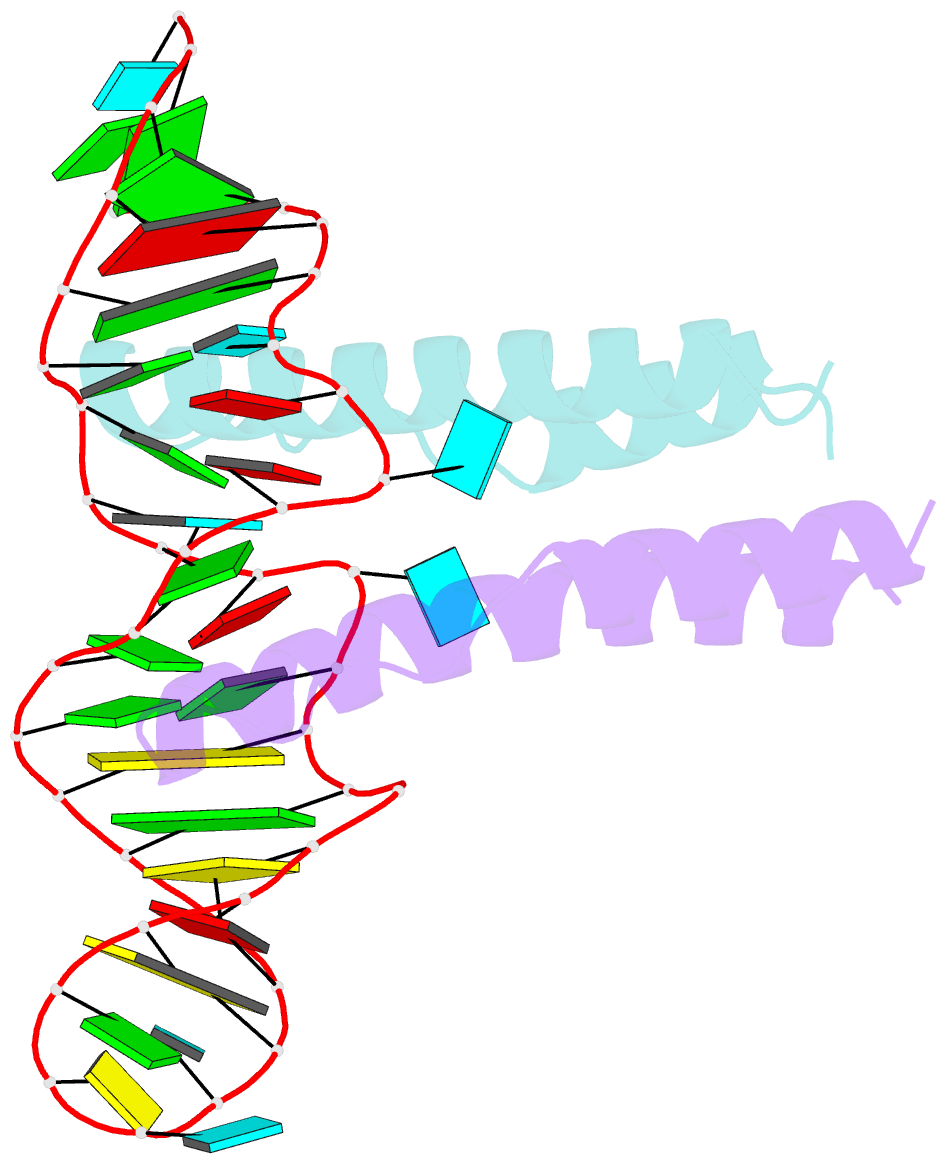
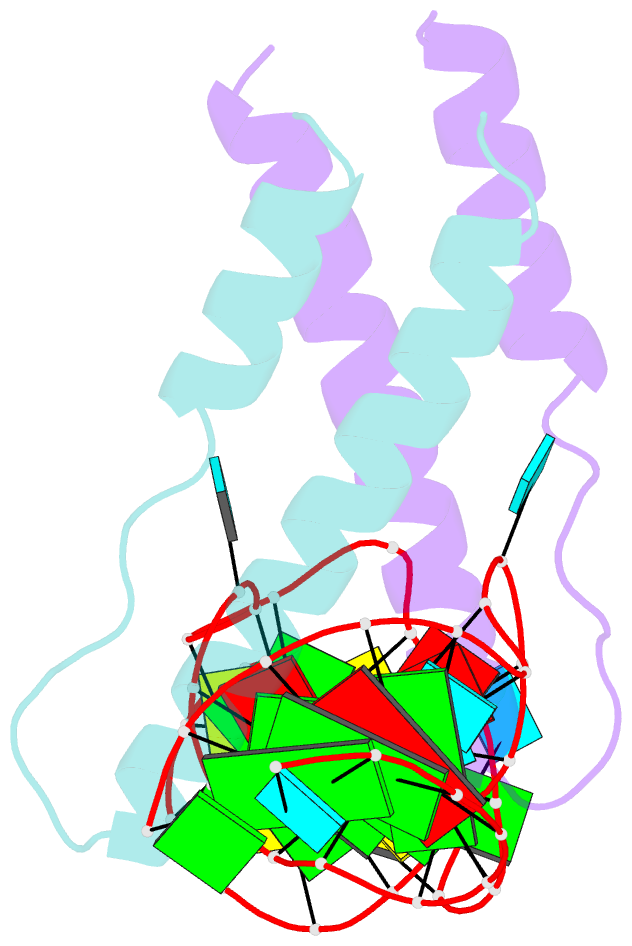
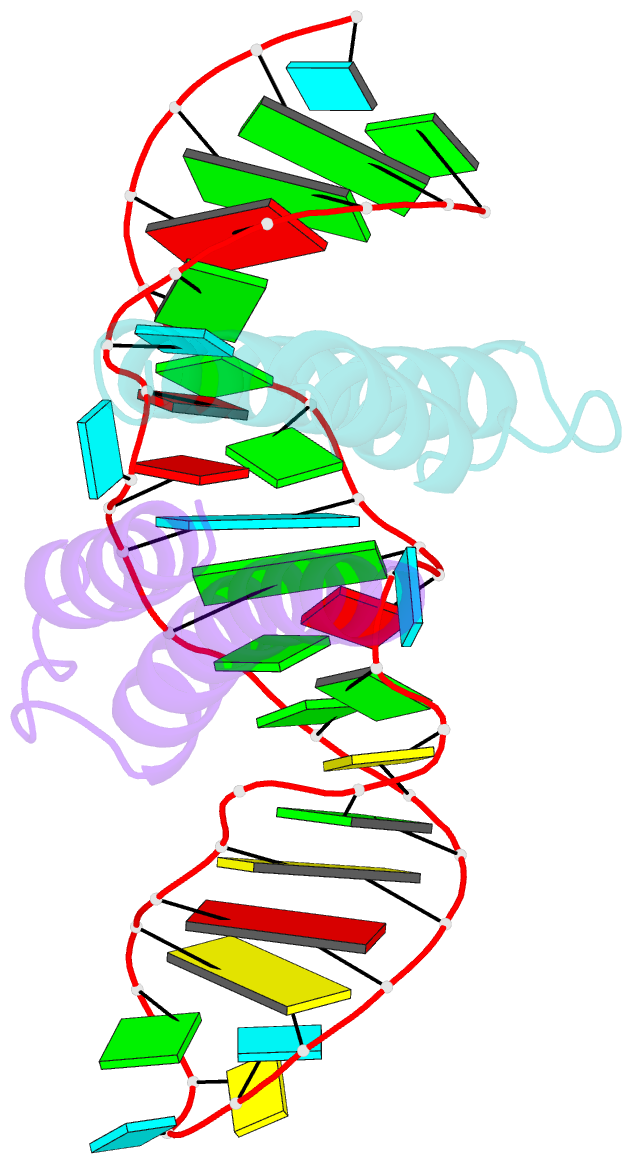
Summary information and primary citation
- PDB-id
- 4pmi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.2 Å)
- Summary
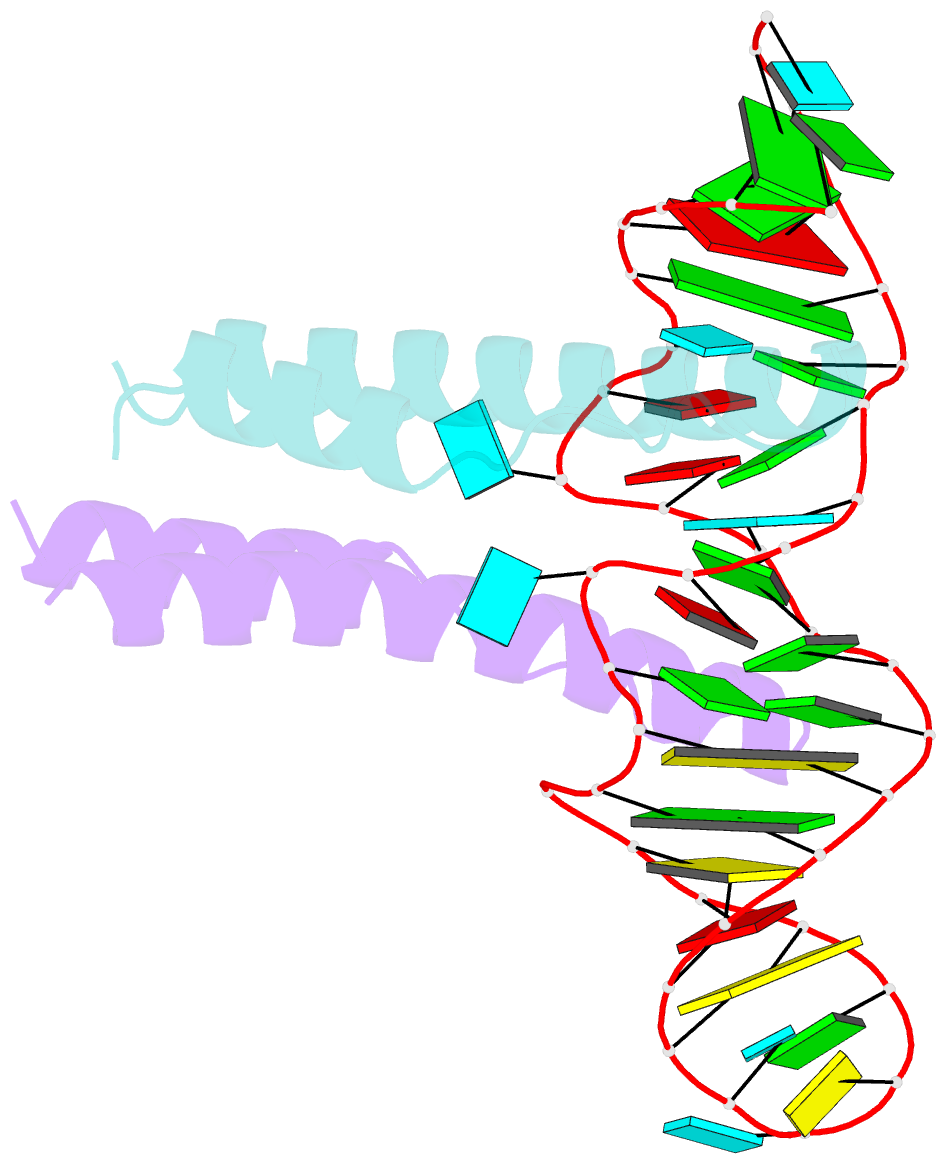
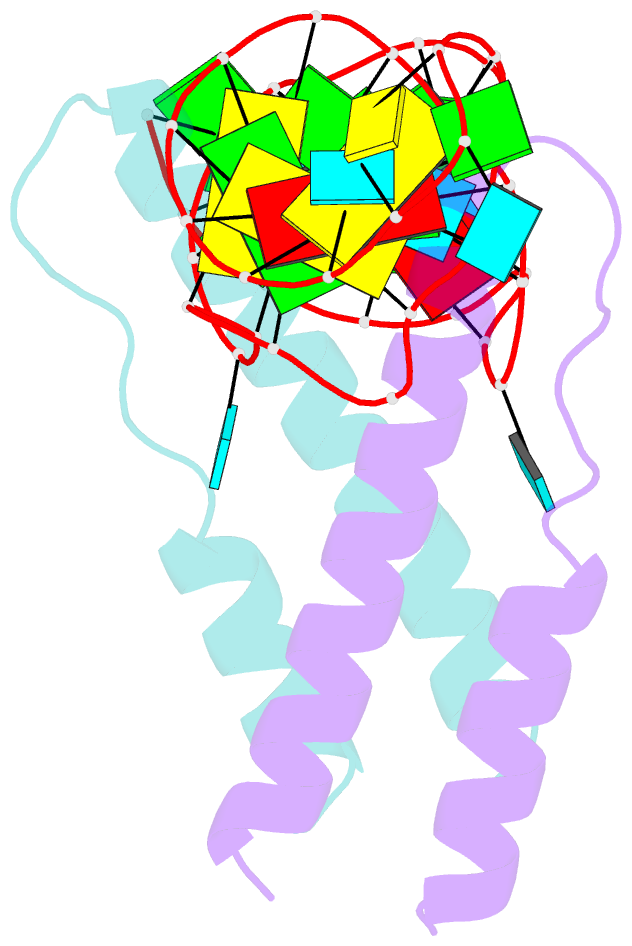
- Crystal structure of rev and rev-response-element RNA complex
- Reference
- Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD (2014): "RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex." Elife, 4, e04120. doi: 10.7554/eLife.04120.
- Abstract
- The HIV-1 protein Rev controls a critical step in viral replication by mediating the nuclear export of unspliced and singly-spliced viral mRNAs. Multiple Rev subunits assemble on the Rev Response Element (RRE), a structured region present in these RNAs, and direct their export through the Crm1 pathway. Rev-RRE assembly occurs via several Rev oligomerization and RNA-binding steps, but how these steps are coordinated to form an export-competent complex is unclear. Here, we report the first crystal structure of a Rev dimer-RRE complex, revealing a dramatic rearrangement of the Rev-dimer upon RRE binding through re-packing of its hydrophobic protein-protein interface. Rev-RNA recognition relies on sequence-specific contacts at the well-characterized IIB site and local RNA architecture at the second site. The structure supports a model in which the RRE utilizes the inherent plasticity of Rev subunit interfaces to guide the formation of a functional complex.