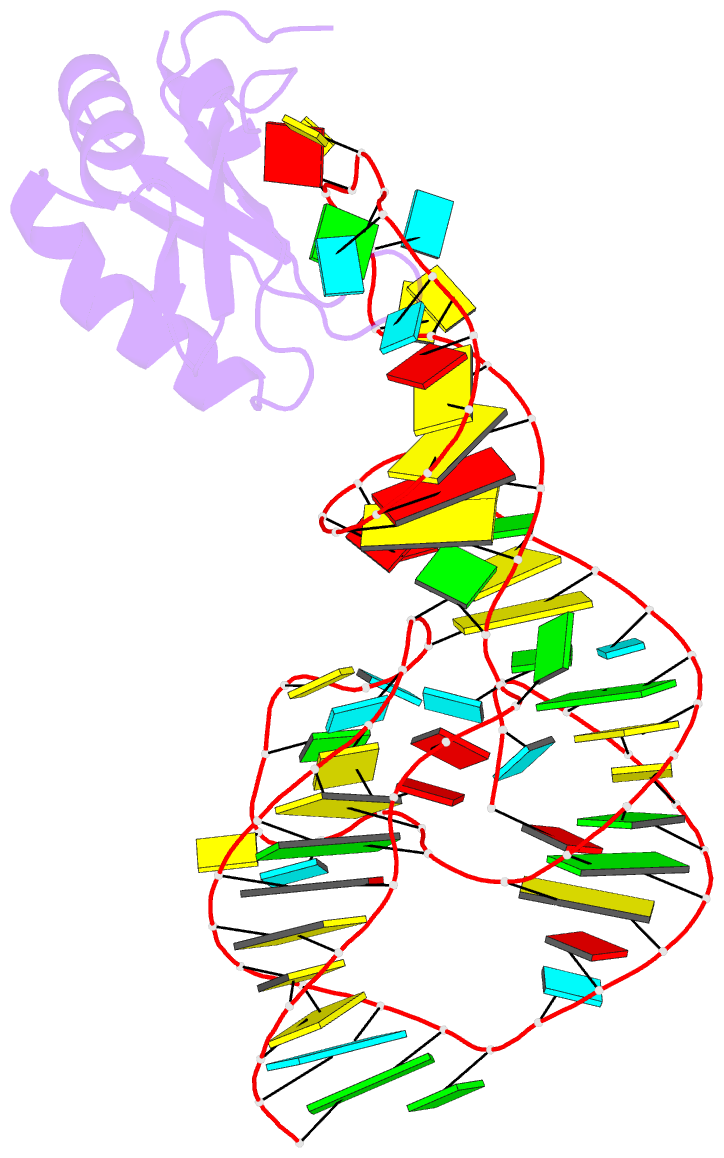
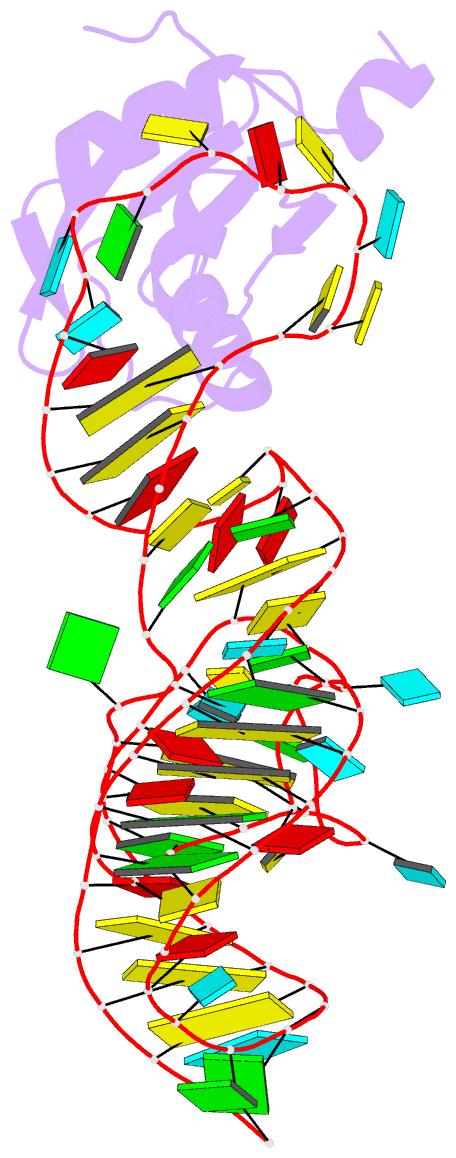
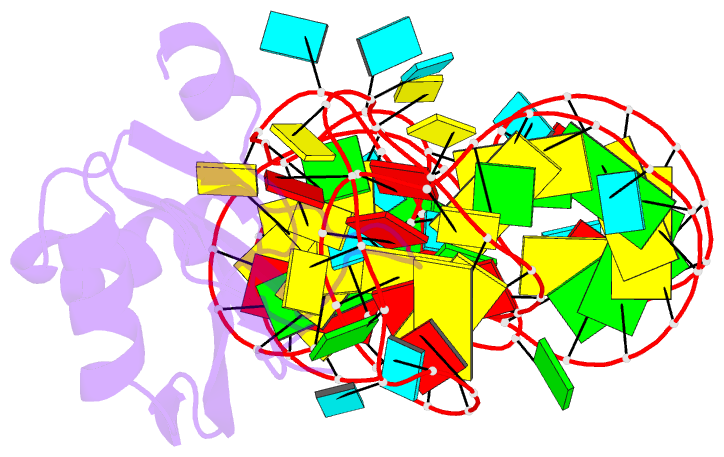
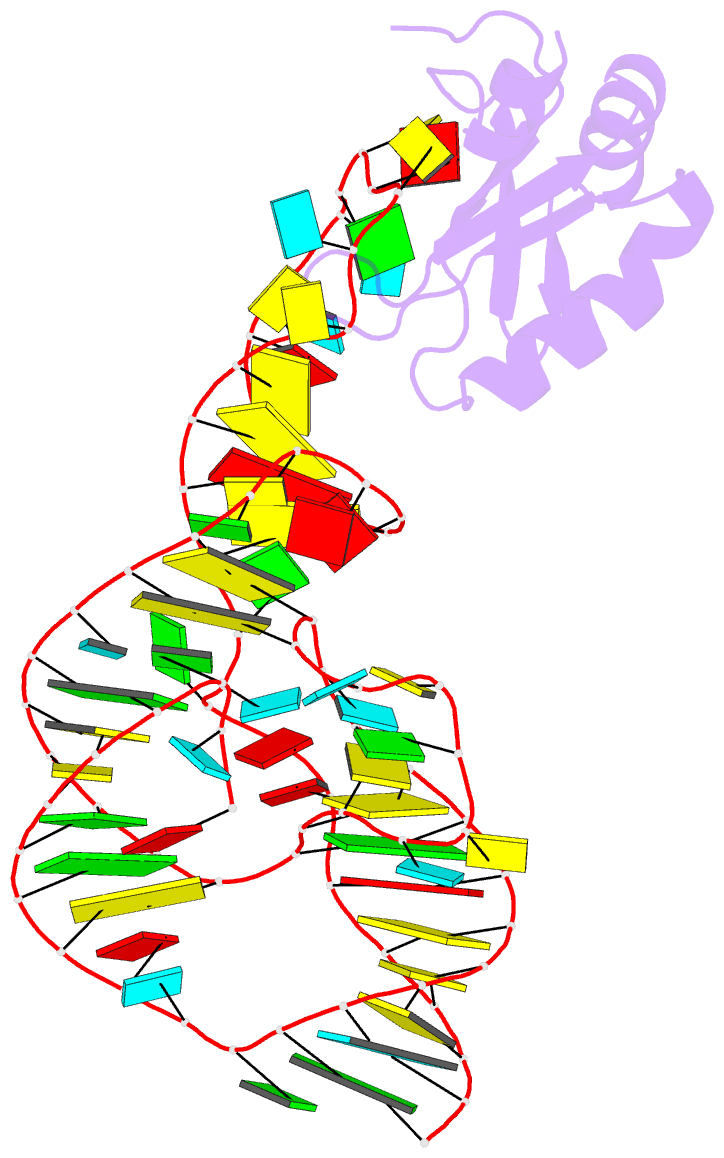
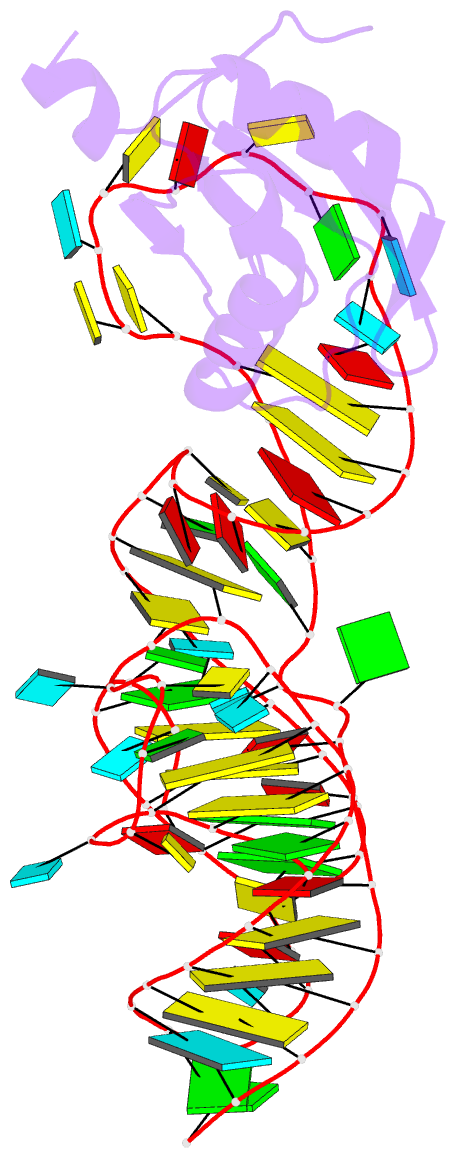
Summary information and primary citation
- PDB-id
-
4prf;
DSSR-derived features in text and
JSON formats; DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.395 Å)
- Summary
- A second look at the hdv ribozyme structure and
dynamics.
- Reference
-
Kapral GJ, Jain S, Noeske J, Doudna JA, Richardson DC,
Richardson JS (2014): "New tools
provide a second look at HDV ribozyme structure, dynamics
and cleavage." Nucleic Acids Res.,
42, 12833-12846. doi: 10.1093/nar/gku992.
- Abstract
- The hepatitis delta virus (HDV) ribozyme is a
self-cleaving RNA enzyme essential for processing viral
transcripts during rolling circle viral replication. The
first crystal structure of the cleaved ribozyme was solved
in 1998, followed by structures of uncleaved,
mutant-inhibited and ion-complexed forms. Recently, methods
have been developed that make the task of modeling RNA
structure and dynamics significantly easier and more
reliable. We have used ERRASER and PHENIX to rebuild and
re-refine the cleaved and cis-acting C75U-inhibited
structures of the HDV ribozyme. The results correct local
conformations and identify alternates for RNA residues,
many in functionally important regions, leading to improved
R values and model validation statistics for both
structures. We compare the rebuilt structures to a higher
resolution, trans-acting deoxy-inhibited structure of the
ribozyme, and conclude that although both inhibited
structures are consistent with the currently accepted
hammerhead-like mechanism of cleavage, they do not add
direct structural evidence to the biochemical and modeling
data. However, the rebuilt structures (PDBs: 4PR6, 4PRF)
provide a more robust starting point for research on the
dynamics and catalytic mechanism of the HDV ribozyme and
demonstrate the power of new techniques to make significant
improvements in RNA structures that impact biologically
relevant conclusions.