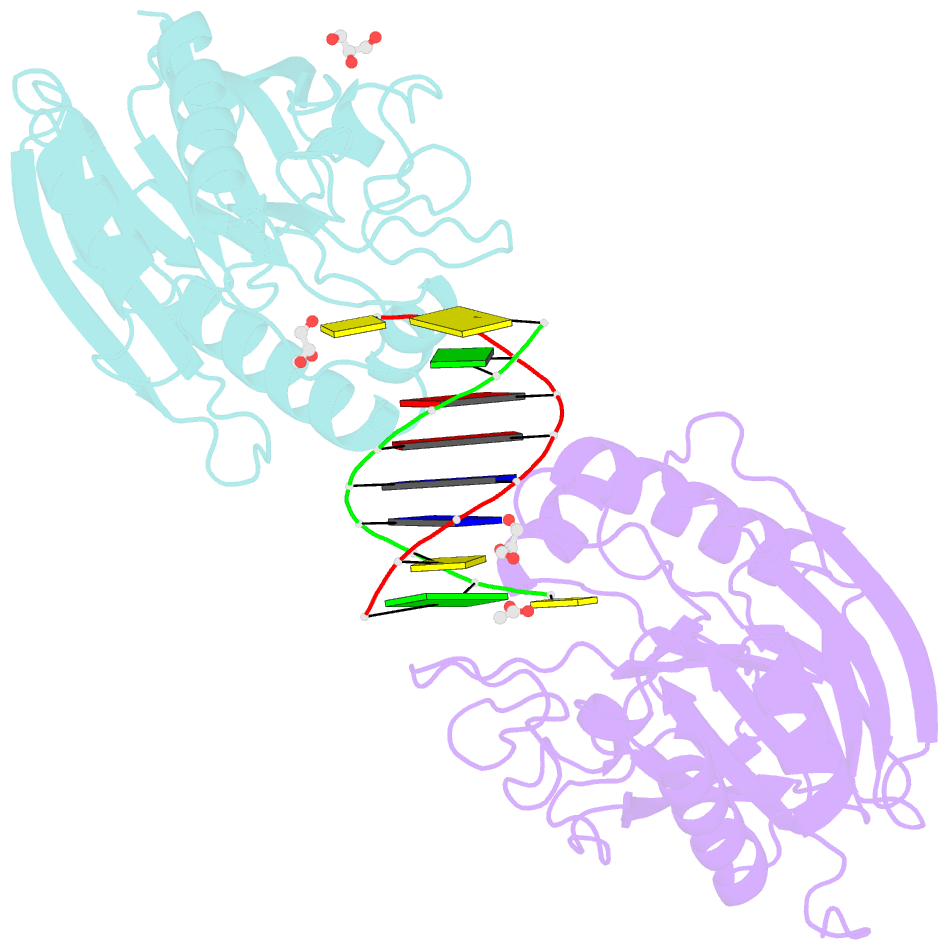
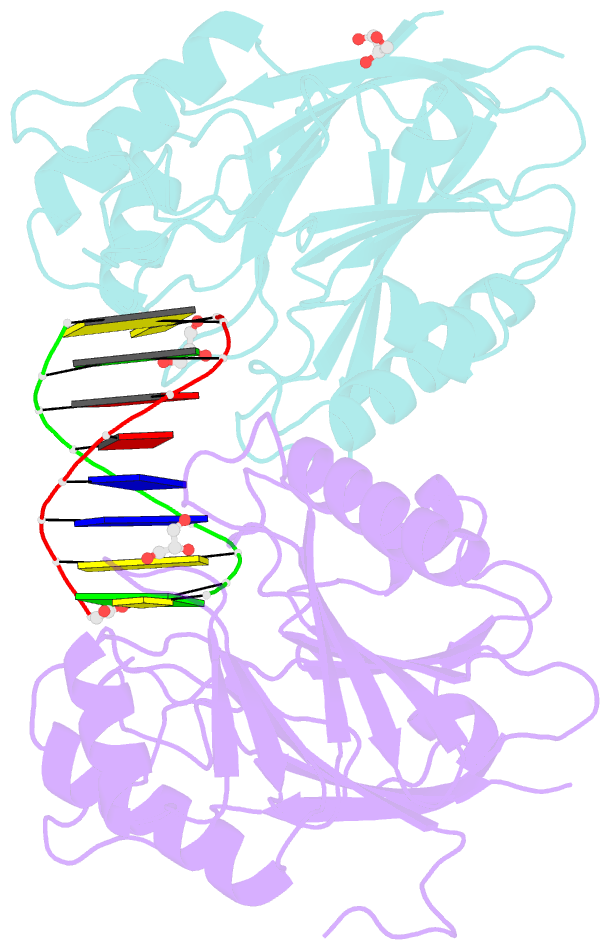
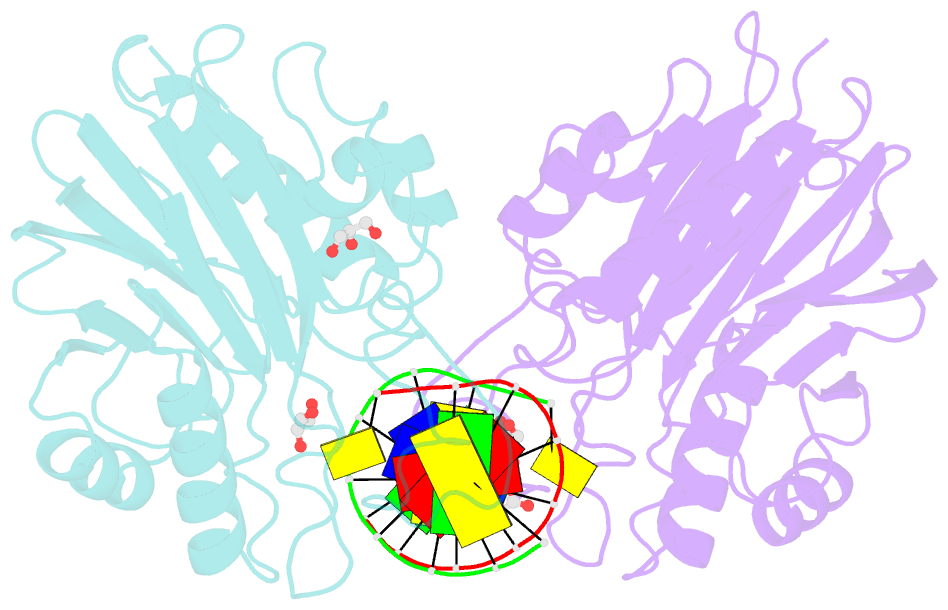
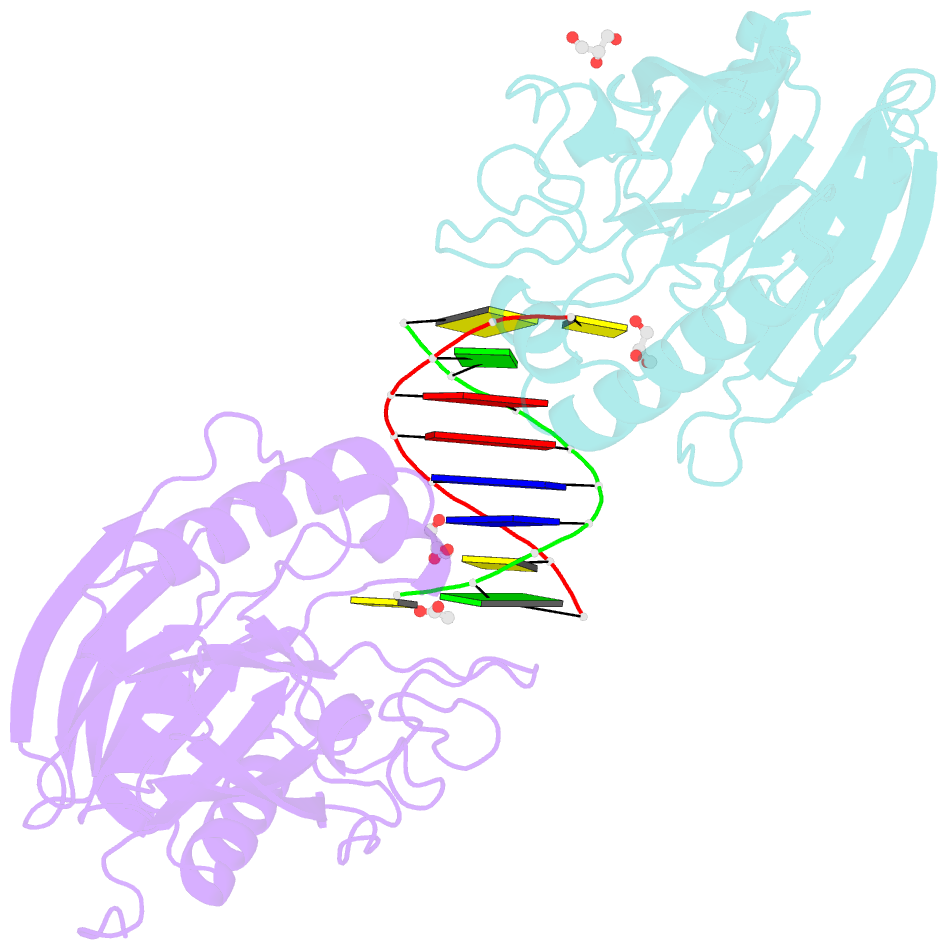
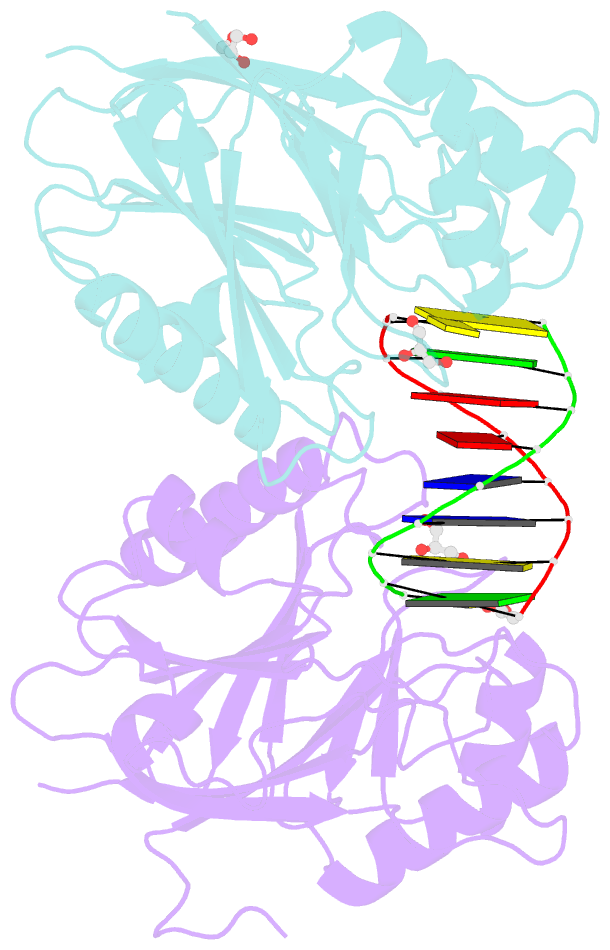
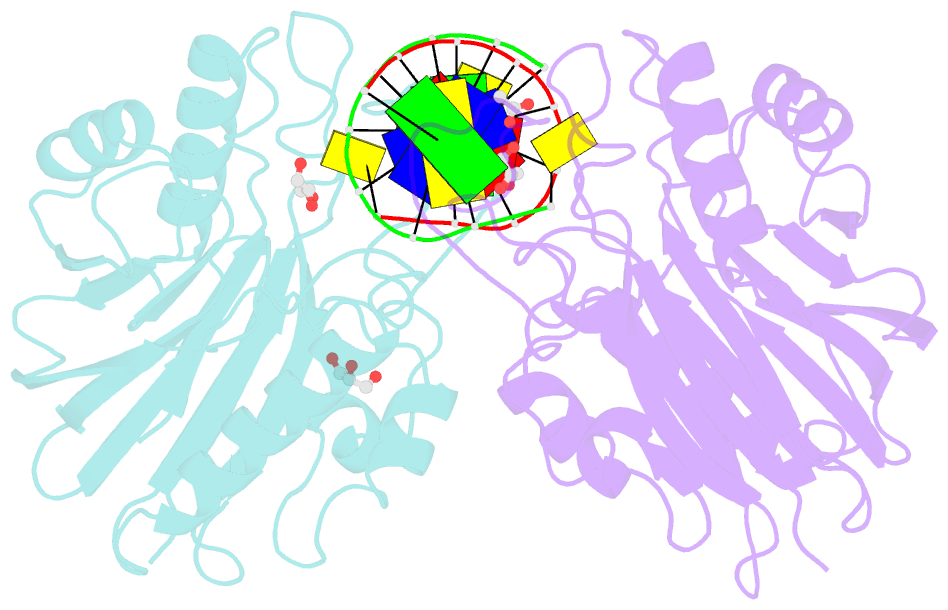
Summary information and primary citation
- PDB-id
- 4puq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA-RNA
- Method
- X-ray (1.6 Å)
- Summary
- Mus musculus tdp2 reaction product complex with 5'-phosphorylated RNA-DNA, glycerol, and mg2+
- Reference
- Gao R, Schellenberg MJ, Huang SY, Abdelmalak M, Marchand C, Nitiss KC, Nitiss JL, Williams RS, Pommier Y (2014): "Proteolytic Degradation of Topoisomerase II (Top2) Enables the Processing of Top2DNA and Top2RNA Covalent Complexes by Tyrosyl-DNA-Phosphodiesterase 2 (TDP2)." J.Biol.Chem., 289, 17960-17969. doi: 10.1074/jbc.M114.565374.
- Abstract
- Eukaryotic type II topoisomerases (Top2α and Top2β) are homodimeric enzymes; they are essential for altering DNA topology by the formation of normally transient double strand DNA cleavage. Anticancer drugs (etoposide, doxorubicin, and mitoxantrone) and also Top2 oxidation and DNA helical alterations cause potentially irreversible Top2·DNA cleavage complexes (Top2cc), leading to Top2-linked DNA breaks. Top2cc are the therapeutic mechanism for killing cancer cells. Yet Top2cc can also generate recombination, translocations, and apoptosis in normal cells. The Top2 protein-DNA covalent complexes are excised (in part) by tyrosyl-DNA-phosphodiesterase 2 (TDP2/TTRAP/EAP2/VPg unlinkase). In this study, we show that irreversible Top2cc induced in suicidal substrates are not processed by TDP2 unless they first undergo proteolytic processing or denaturation. We also demonstrate that TDP2 is most efficient when the DNA attached to the tyrosyl is in a single-stranded configuration and that TDP2 can efficiently remove a tyrosine linked to a single misincorporated ribonucleotide or to polyribonucleotides, which expands the TDP2 catalytic profile with RNA substrates. The 1.6-Å resolution crystal structure of TDP2 bound to a substrate bearing a 5'-ribonucleotide defines a mechanism through which RNA can be accommodated in the TDP2 active site, albeit in a strained conformation.