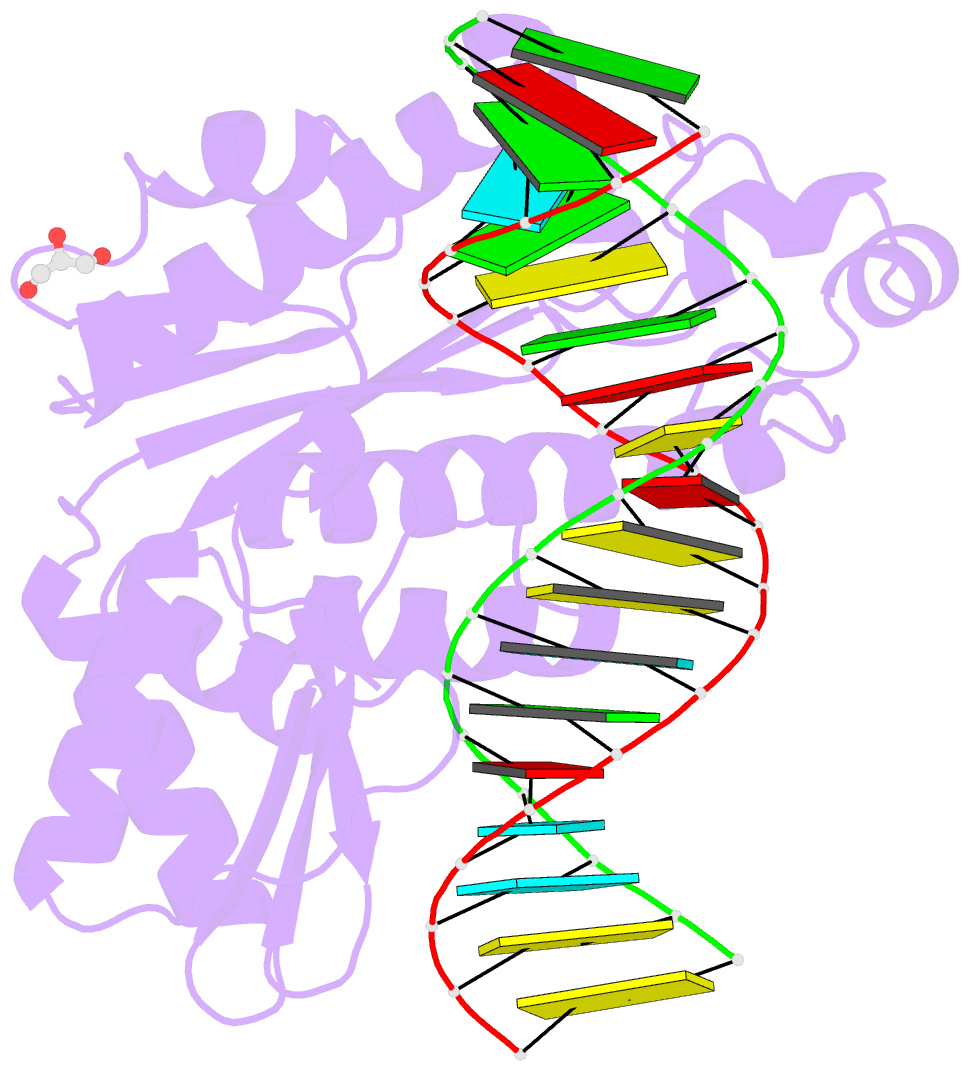
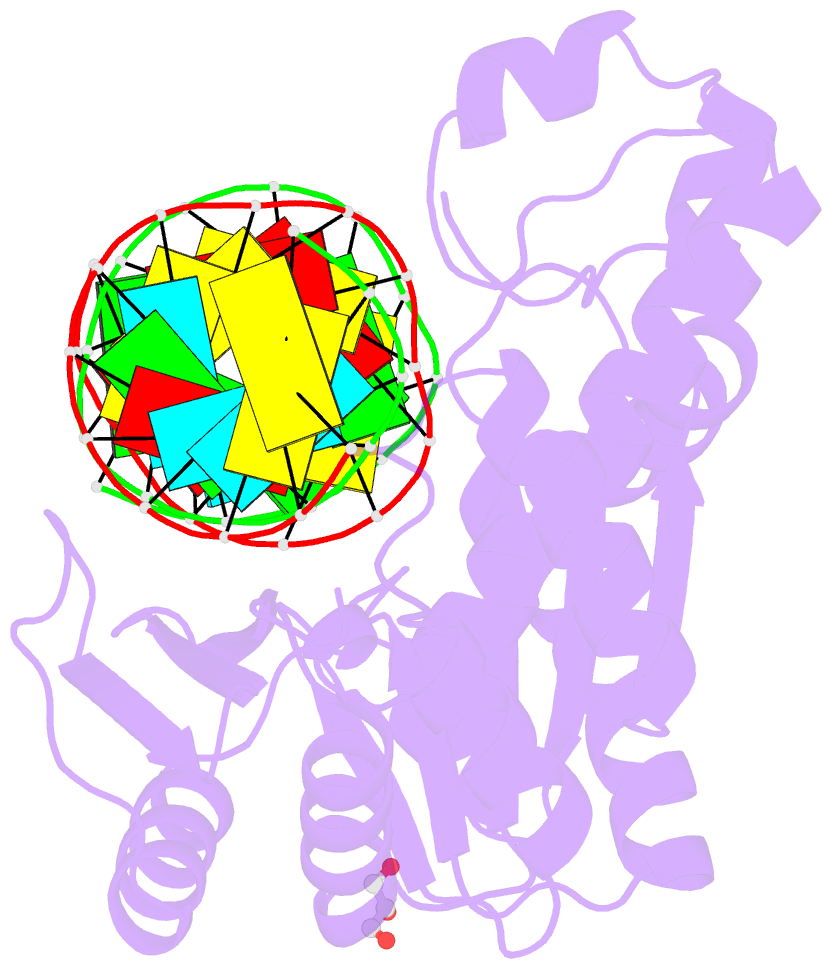
Summary information and primary citation
- PDB-id
- 4py5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA-RNA
- Method
- X-ray (2.1 Å)
- Summary
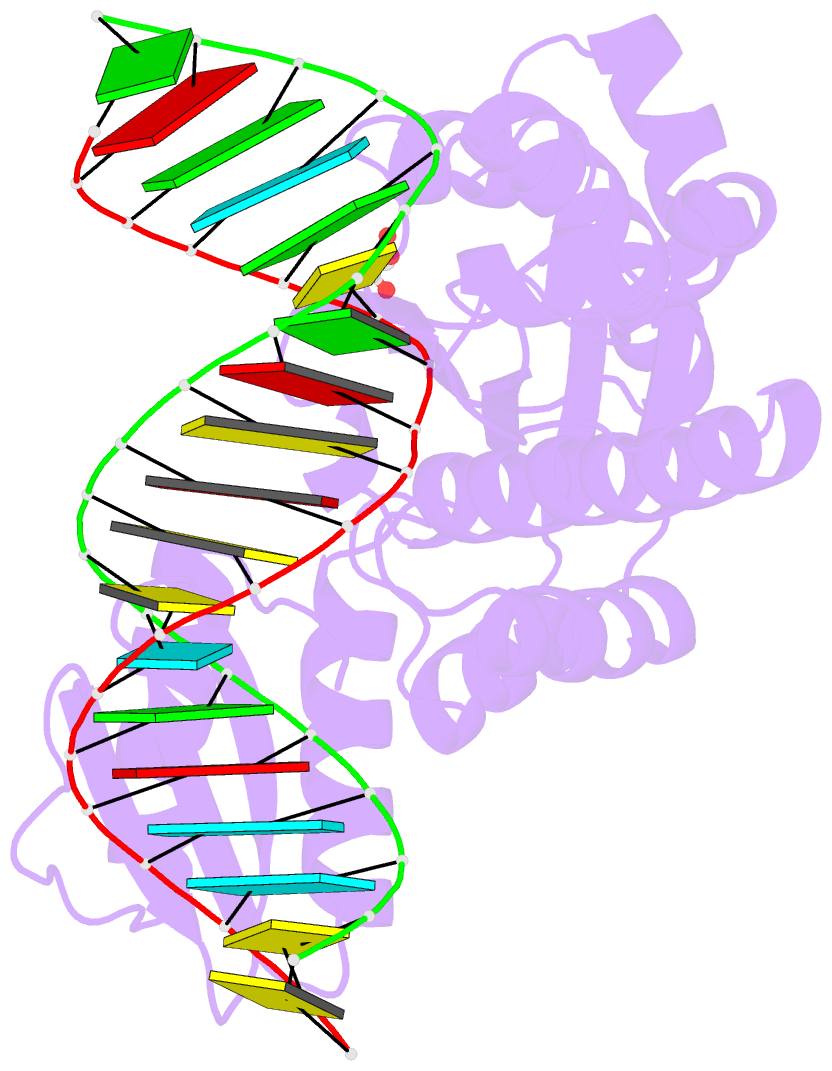
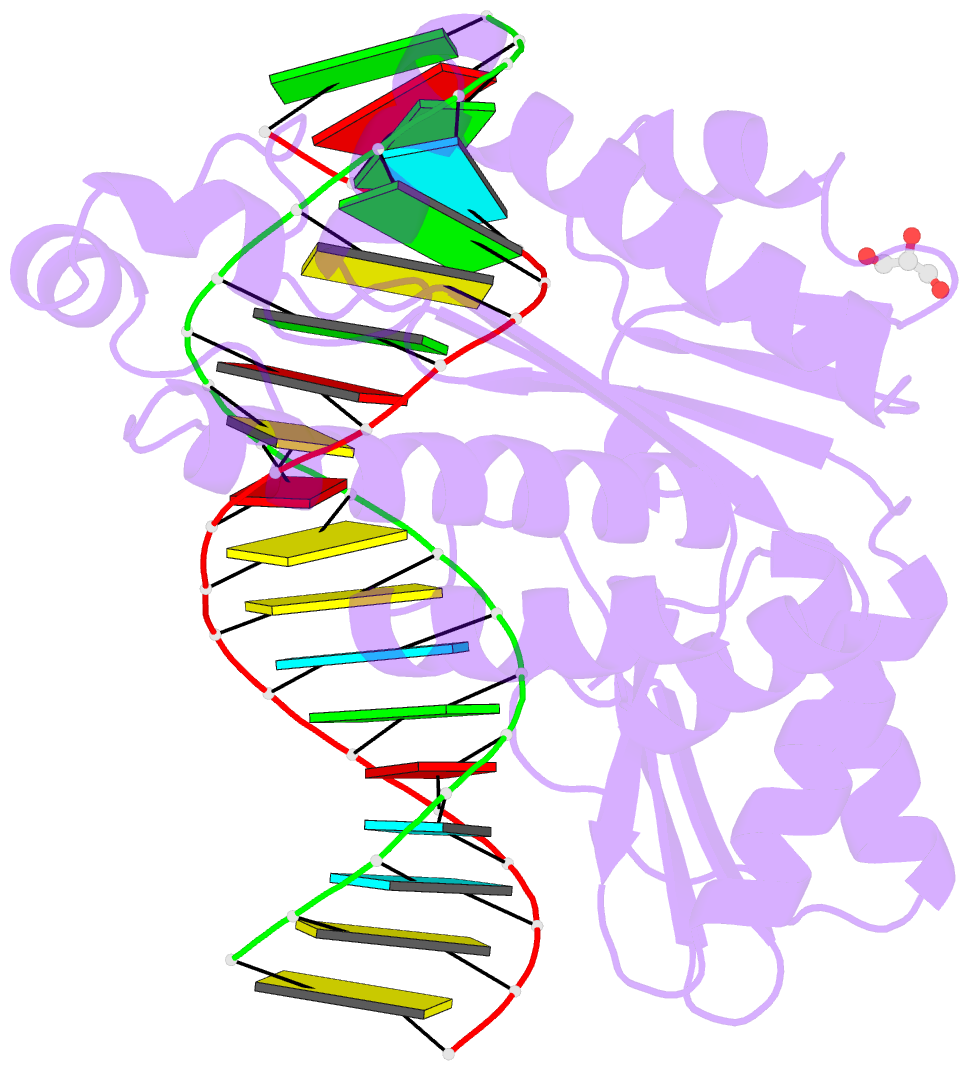
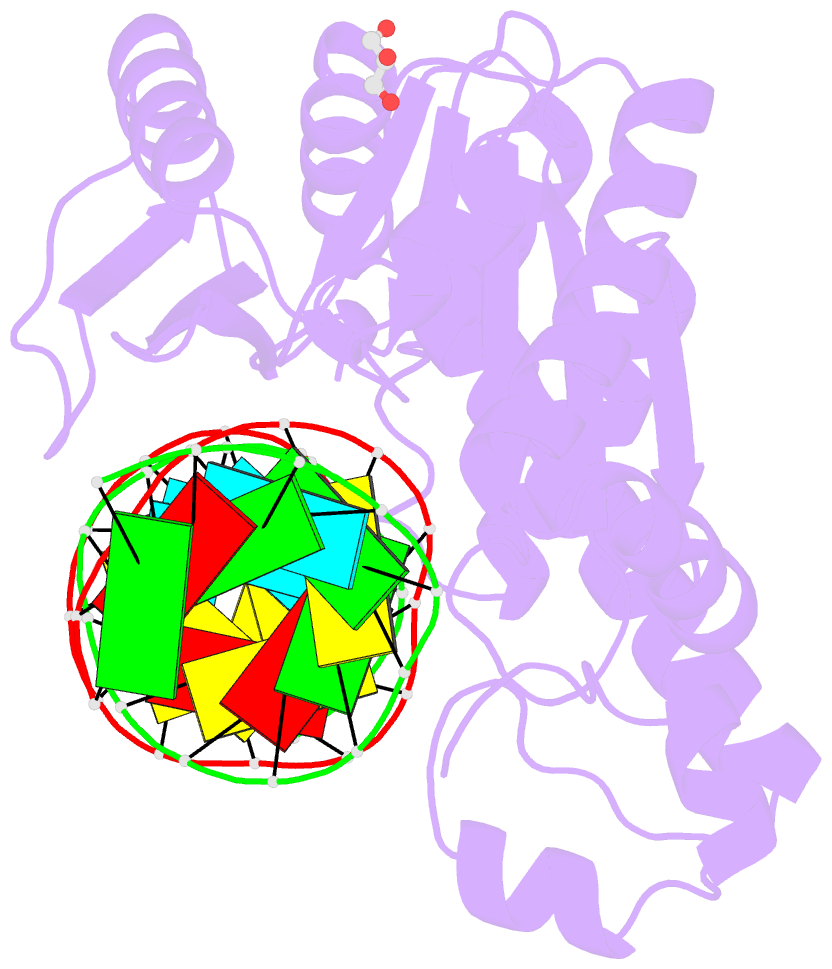
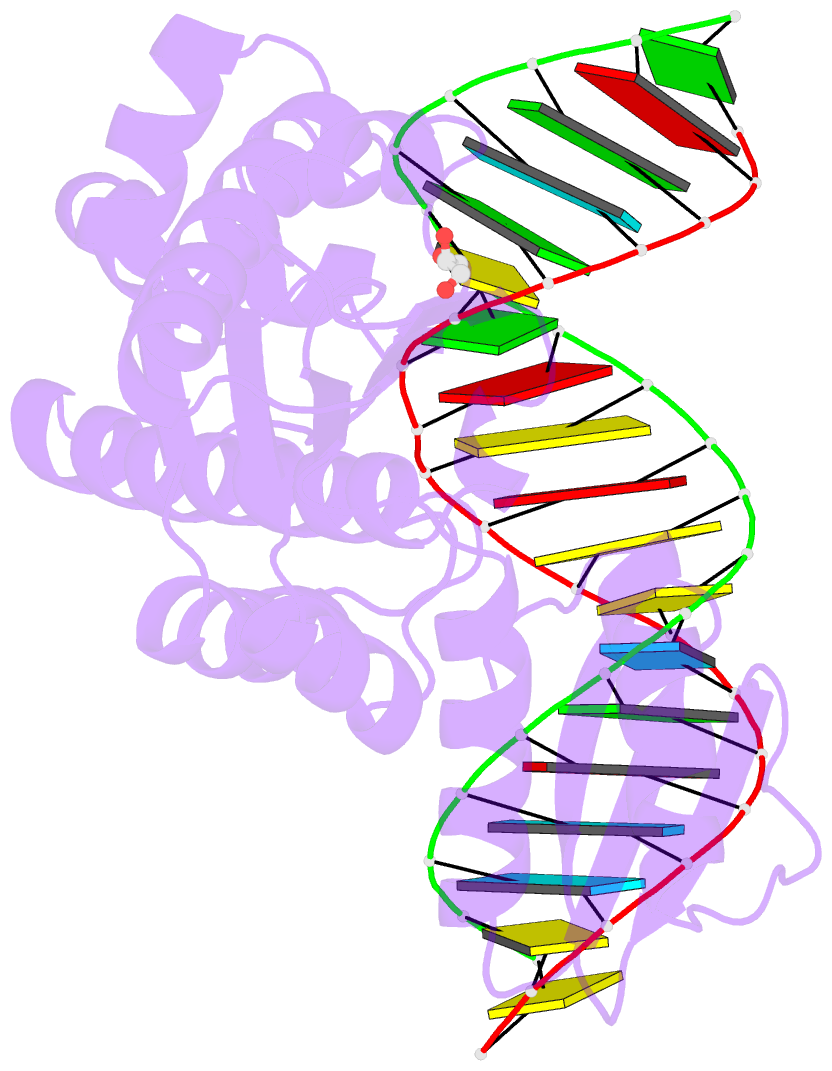
- Thermovibrio ammonificans rnase h3 in complex with 19-mer RNA-DNA
- Reference
- Figiel M, Nowotny M (2014): "Crystal structure of RNase H3-substrate complex reveals parallel evolution of RNA/DNA hybrid recognition." Nucleic Acids Res., 42, 9285-9294. doi: 10.1093/nar/gku615.
- Abstract
- RNases H participate in the replication and maintenance of genomic DNA. RNase H1 cleaves the RNA strand of RNA/DNA hybrids, and RNase H2 in addition hydrolyzes the RNA residue of RNA-DNA junctions. RNase H3 is structurally closely related to RNases H2, but its biochemical properties are similar to type 1 enzymes. Its unique N-terminal substrate-binding domain (N-domain) is related to TATA-binding protein. Here, we report the first crystal structure of RNase H3 in complex with its RNA/DNA substrate. Just like RNases H1, type 3 enzyme recognizes the 2'-OH groups of the RNA strand and detects the DNA strand by binding a phosphate group and inducing B-form conformation. Moreover, the N-domain recognizes RNA and DNA in a manner that is highly similar to the hybrid-binding domain of RNases H1. Our structure demonstrates a remarkable example of parallel evolution of the elements used in the specific recognition of RNA and DNA.