Summary information and primary citation
- PDB-id
- 4qm6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (1.5 Å)
- Summary
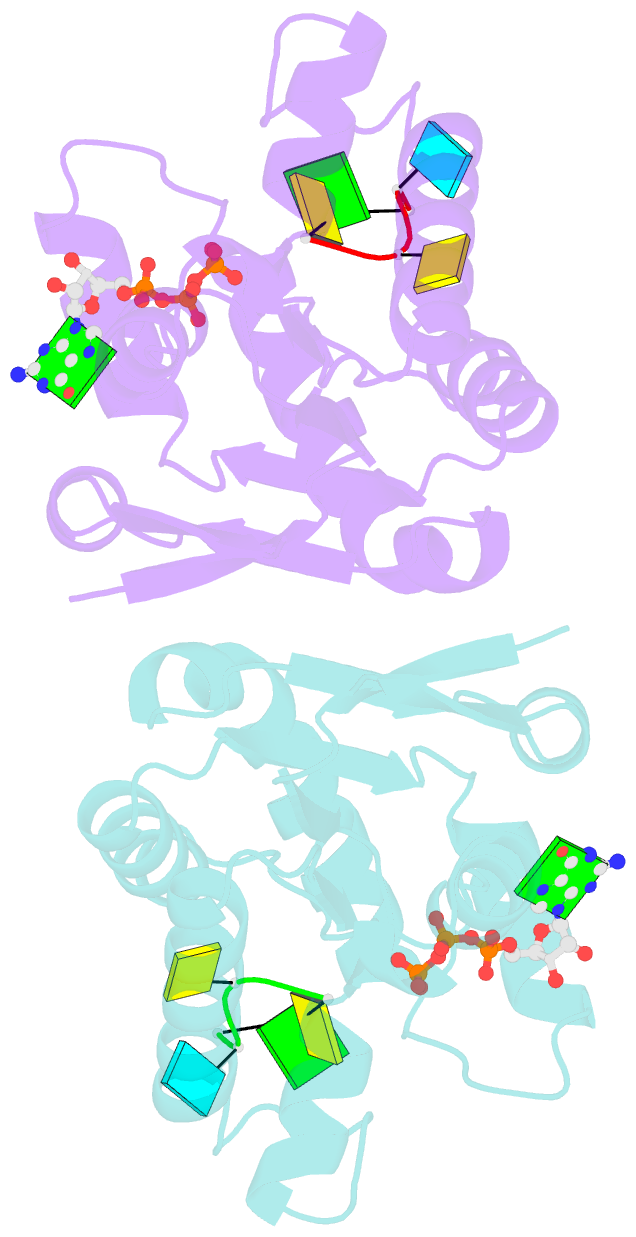
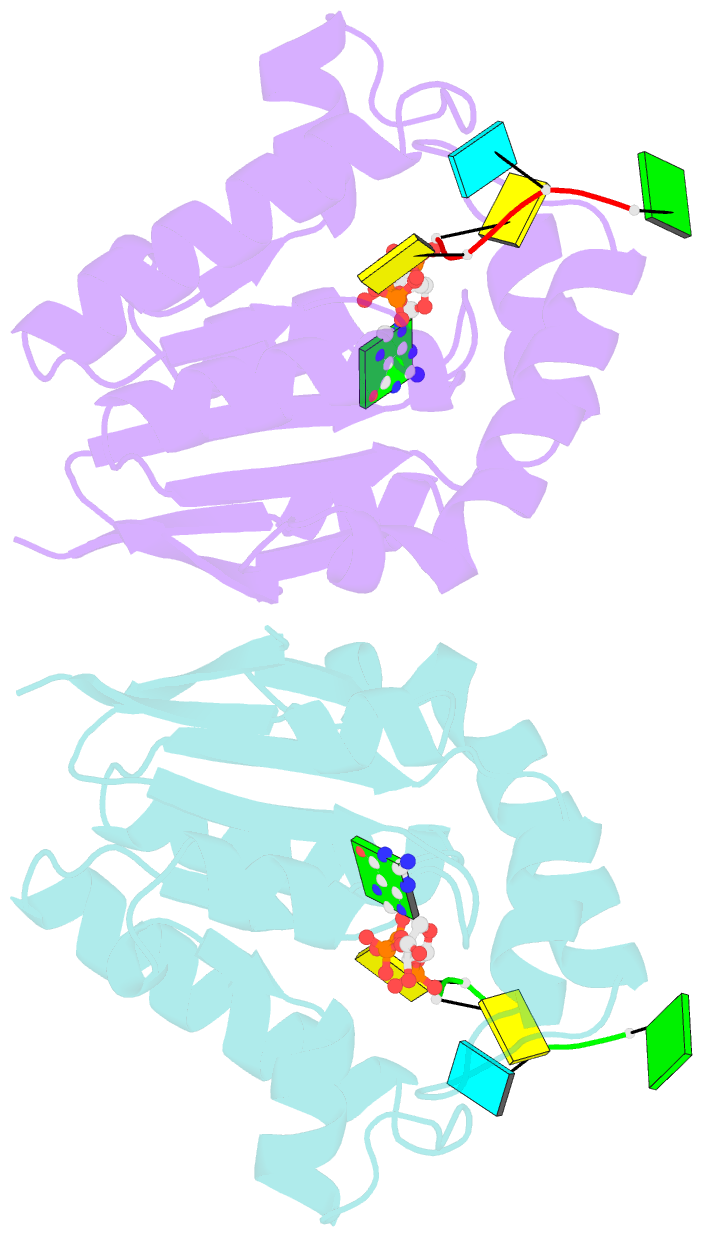
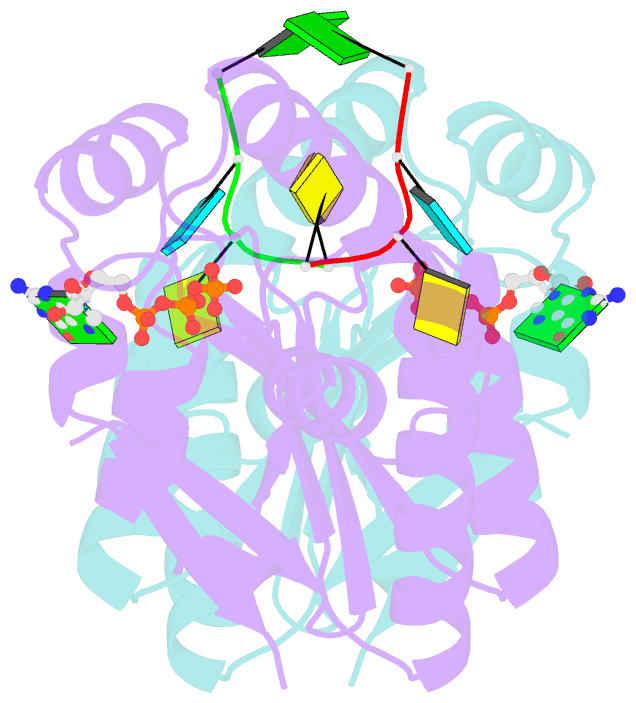
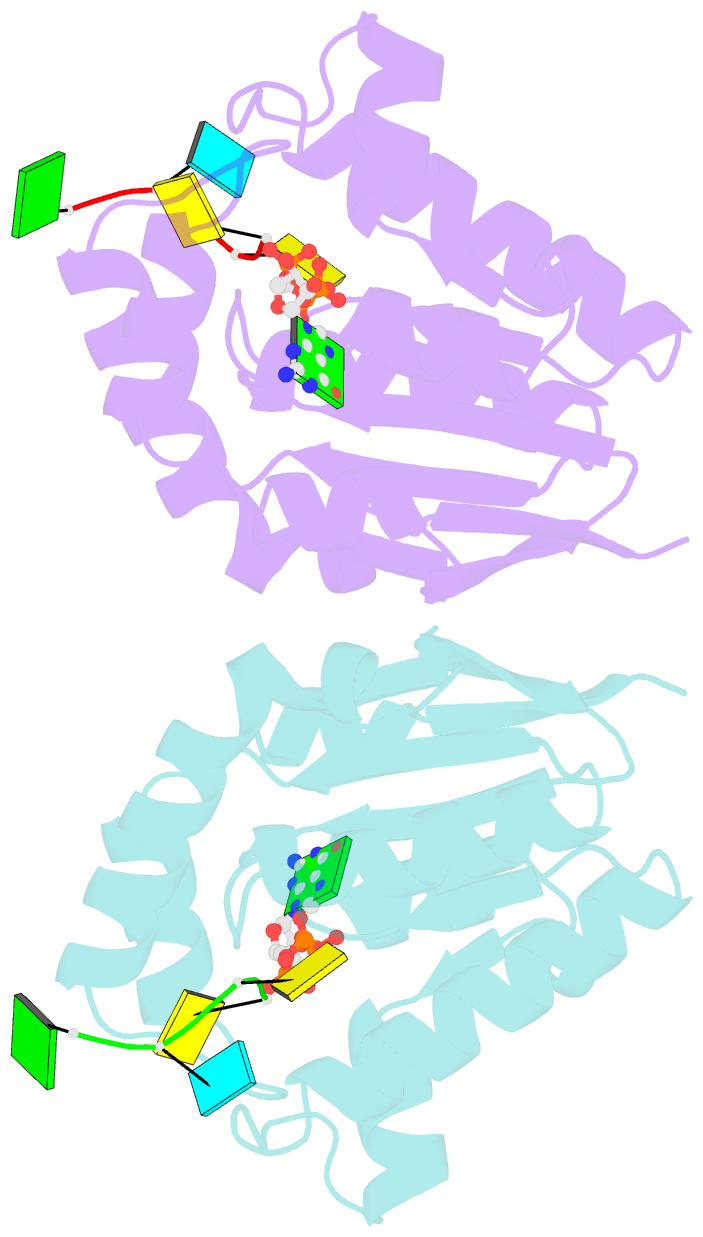
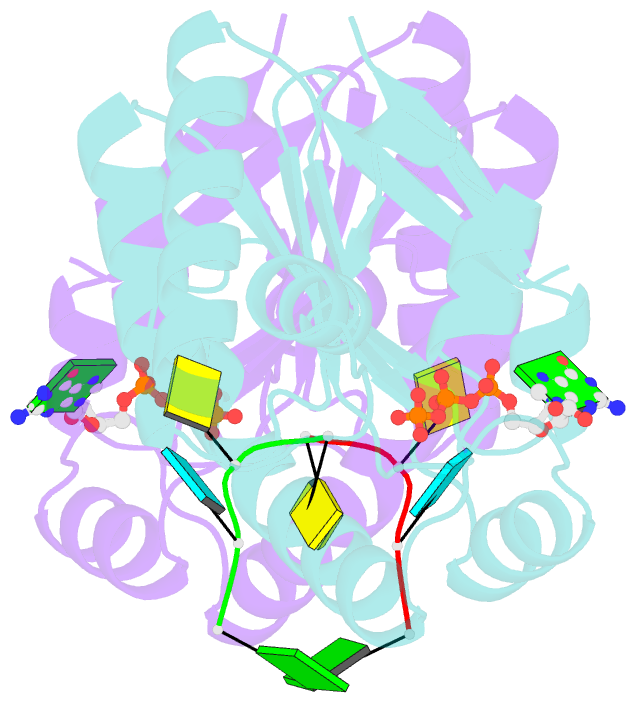
- Structure of bacterial polynucleotide kinase bound to gtp and RNA
- Reference
- Das U, Wang LK, Smith P, Munir A, Shuman S (2014): "Structures of bacterial polynucleotide kinase in a michaelis complex with nucleoside triphosphate (NTP)-Mg2+ and 5'-OH RNA and a mixed substrate-product complex with NTP-Mg2+ and a 5'-phosphorylated oligonucleotide." J.Bacteriol., 196, 4285-4292. doi: 10.1128/JB.02197-14.
- Abstract
- Clostridium thermocellum polynucleotide kinase (CthPnk), the 5'-end-healing module of a bacterial RNA repair system, catalyzes reversible phosphoryl transfer from a nucleoside triphosphate (NTP) donor to a 5'-OH polynucleotide acceptor, either DNA or RNA. Here we report the 1.5-Å crystal structure of CthPnk-D38N in a Michaelis complex with GTP-Mg(2+) and a 5'-OH RNA oligonucleotide. The RNA-binding mode of CthPnk is different from that of the metazoan RNA kinase Clp1. CthPnk makes hydrogen bonds to the ribose 2'-hydroxyls of the 5' terminal nucleoside, via Gln51, and the penultimate nucleoside, via Gln83. The 5'-terminal nucleobase is sandwiched by Gln51 and Val129. Mutating Gln51 or Val129 to alanine reduced kinase specific activity 3-fold. Ser37 and Thr80 donate functionally redundant hydrogen bonds to the terminal phosphodiester; a S37A-T80A double mutation reduced kinase activity 50-fold. Crystallization of catalytically active CthPnk with GTP-Mg(2+) and a 5'-OH DNA yielded a mixed substrate-product complex with GTP-Mg(2+) and 5'-PO4 DNA, wherein the product 5' phosphate group is displaced by the NTP γ phosphate and the local architecture of the acceptor site is perturbed.